Vol-7, Special Issue3-April, 2016, pp957-966 http://www.bipublication.com
Case Report
Functional Exposed Amino Acids of OSPA as a
Candidate for Lyme disease Vaccine
Negin Kafee1 and Fateme Sefid2*
1Departement of Biology, Science and Art University,
Yazd, Iran Email:negin.kafee@yahoo.com
*2Department of Biology, shahed University ,
Tehran-Qom Express Way E-mail: sefid@shahed.ac.ir
*
Corresponding author: Biology Department, Shahed University, Tehran-Qom Express way, Iran. Email: sefid@shahed.ac.ir and sefid.fateme@yahoo.com
ABSTRACT:
Lyme disease is caused by the bacterium Borreliaburgdorferi and is transmitted to humans through the bite of infected black legged ticks. Typical symptoms include fever, headache, fatigue, and a characteristic skin rash called erythema migrans. If left untreated, infection can spread to joints, the heart, and the nervous system. Lyme disease is diagnosed based on symptoms, physical findings (e.g., rash), and the possibility of exposure to infected ticks. The protective role of antibodies to B. burgdorferi has been explored in animal models of Lyme disease. Findings indicated that antibodies against OspA were protective, making this antigen a likely candidate for a vaccine. Early clinical trials demonstrated that recombinant OspA was immunogenic and well tolerated, even in subjects with a history of Lyme disease. The present study was designed to in silico resolving the major obstacles in the control or in prevention of lyme diseases. We exploited bioinformatic tools to better understanding and characterizing the OspAstructure and select appropriate regions as effectiveB cell epitops.
Keywords: B cell epitops, Borreliaburgdorferi, outer surface proteins
INTRODUCTION:
The spirochete Borreliaburgdorferi is a tick-borne obligate parasite whose normal reservoir is a variety of small mammals[1]. Whereas infection of these natural hosts does not lead to disease, infection of humans can result in Lyme disease, as a consequence of the human immuno pathological response to B. burgdorferi[2]. Consistent with the pathogenesis of Lyme disease, bacterial products that allow B. burgdorferi to replicate and survive, rather than true “virulence factors,” appear to be primarily what is required for the bacterium to cause disease in a susceptible host[3-8].Lyme
disease is caused by the bacterium
(Osp) that have been characterized (OspA through OspF) [18]. The Osp proteins are lipoproteins anchored by N-terminally attached fatty acid molecules to the membrane[19].
They are presumed to play a role in virulence, transmission, or survival in the tick. B. burgdorferi elicits an antibody response to a repertoire of borrelial proteins, including outer surface protein C (OspC) and flagellin. In the early stage of the disease, free antibodies to OspC and flagellin are measurable, whereas only later are responses to outer-surface protein A (OspA a surface lipoprotein demonstrable in standard assays[20-22].A number of studies have implicated OspA as a protective antigen and OspA-specific antibodies as protective against Lyme borreliosis in the mouse model[23].
Passive transfer of monoclonal and polyclonal sera specific for OspA was shown to protect mice against challenge with tgburgdoq'eri. Removal of antibodies specific for OspA greatly diminished the protective component in polyclonal sera generated by immunization with whole killed Borrelia[24].
Studies using non-lipid acylatedOspA-glutathione S-transferase (OspA-GST) fusion protein in active immunizations have also demonstrated promising protection in the mouse model for Lyme borreliosis; however, protection required repeated immunizations with large doses of OspAGST and formulation with CFA, which is unsuitable for human use. As a lipoprotein, OspA is insoluble and not expressed well in E. coli; accordingly, OspA can be unwieldy for immunogenicity studies in the laboratory or large-scale production as a subunit vaccine[25]. Recently, however, it was demonstrated that purified OspA lipoprotein (L-OspA) is profoundly more immunogenic than nonlipidacylatedOspA (NL-OspA), even in the absence of adjuvant[26].
The protective role of antibodies to B. burgdorferi has been explored in animal models of Lyme disease. Findings indicated that antibodies against OspA were protective, making this antigen a likely candidate for a vaccine.
Early clinical trials demonstrated that recombinant OspA was immunogenic and well tolerated, even in subjects with a history of Lyme disease[27,28]. A dose-ranging study showed that as compared with doses of 1, 5, and 10 µg, a dose of 30 µg of the OspA vaccine elicited an optimal antibody response without causing an increased rate of symptomatic reactions[29].
The property of an antigen to bind specifically complementary antibodies is known as the antigen’s antigenicity; likewise, the ability of an antigen to induce an immune response is called its immunogenicity. Attempts should be made to discover peptides that could mimic protein epitopes and possess the same immunogenicity as the whole protein. Subsequently, theoretical methods for epitope prediction have been developed leading to synthesis of such peptides
that are important for development of
immunodiagnostic tests and vaccines.
The present study was designed to in silico resolving the major obstacles in the control or in prevention of lyme diseases. We exploited bioinformatic tools to better understanding and characterizing the OspAstructure and select appropriate regions as effectiveB cell epitops.
2.MATERIAL AND METHODS 2.1. Sequence availability
The OspA protein sequence acquired from NCBI
at http://www.ncbi.nlm.nih.gov/protein were
saved in FASTA format for further analyses.
2.2. Homology search and alignment
The sequences served as a query for protein
BLASTat http://blast.ncbi.nlm.nih.gov/Blast.cgi
against non redundant protein database. Probable putative conserved domains of the query protein were also searched for, at the above address.
2.3. Alignment
generations were carried out by CLC sequence analysis 6.1 software.
The BLOSUM substitution matrix was selected with a gap penalty of 10 and a gap extension penalty of 0.1. Multiple sequence alignment in CLC sequence analysis is based on the CLUSTALW program which aligns multiple sequences using a progressive pairwise alignment algorithm.
2.4. Homology modelling
The SWISS-MODEL(Guex& Peitsch, 1997) Workspace at http://swissmodel.expasy.org/ is a web-based integrated service dedicated to protein structure homology modelling.
It assists and guides the user in building protein
homology models at different levels of
complexity.
Building a homology model comprises four main steps: identification of structural template(s), alignment of target sequence and template structure(s), model building, and model quality evaluation. These steps can be repeated until a satisfying modelling result is achieved. Each of the four steps requires specialized software and access to up-to-date protein sequence and structure databases.
2.5. Models evaluations
All 3D models of the proteins built, were
qualititatively estimated by GMQE and
QMEAN4scores.
2.6. Ligand binding site predictions
Cofactor [38] at
http://zhanglab.ccmb.med.umich.edu/COFACTO R/ is a structure-based method for biological
function annotation of protein molecules.
Important amino acid involved in ligand binding site is predicted by this server.
2.7. Identification of functionally and structurally important residues
InterProSurf at http://curie.utmb.edu/pattest9.html predicting functional sites on protein surface using patch analysis was employed. OspA 3D structure determined in a previous study, served as an input file for this server.
2.8. single-scale amino acid properties assay
IEDB(Zhang, Wang, Kim et al., 2008b) at http://tools.immuneepitope.org/tools/bcell/iedb_in put parameters such as hydrophilicity, flexibility, accessibility, turns and antigenic propensity of polypeptide have been correlated with the location of B cell epitopes. This has led to a search for empirical rules that would allow the position of B cell epitopes to be predicted from certain features of the protein sequence.
2.9. B cell epitope prediction
Ellipro(Ponomarenko, Bui, Li et al., 2008) at http://tools.immuneepitope.org/tools/ElliPro/tutori al.jsp predicts linear and discontinuous antibody epitopes.
3. RESULTS AND DISCUSSION
3.1. Sequence availability Homology search and alignment
The protein sequence with 177 residues
(gi|20384687|gb|AAK38346.1|) obtained from NCBI and saved in FASTA format. Protein sequence serving as query for BLAST produced a set of sequences as the highest similar sequence.
3.2. Homology search and alignment
BLAST search revealed numerous hits to the OspA subunit sequence. All hits were of
Borreliaburgdorferi. Putative conserved domains
Fig: 1.Osp A sequenced alignments with CLC program.
3.3. Homology modelling
Building a homology model comprises four main steps: identification of structural template(s), alignment of target sequence and template structure(s), model building, and model quality evaluation. These steps can be repeated until a satisfying modelling result is achieved. Each of the four steps requires specialized software and access to up-to-date protein sequence and
structure databases.Swiss model software
recruited for homology modeling introduced 3 model. All the models were selected for further analyses.
3.4. Models evaluations
The 3D models estimated qualititatively by tow servers revealed that there was a consensus on a single model.QMEAN is a composite scoring function for the estimation of the global and local model quality. QMEAN consisting of four structural descriptors: The local geometry is analysed by a torsion angle potential over three consecutive amino acids. Two pairwise distance-dependent potentials are used to assess all-atom and C-beta interactions. A solvation potential describes the burial status of the residues. The
increase reliability of the quality estimation. The best 3D model is shown in Figure 2.
Fig: 2. OspA 3D structure model.
3.5. Identification of functionally and structurally important residues
Interprosurf annotated functional residues on the 3D structure of OspA.Residues Predicted by
Auto Patch Analysis are residue number78 and79. Results are shown in figure 3.
. Fig: 3. Functional residues on OspA 3D structure.
3.6. ligand binding site predictions
Ligand binding sites determined using
COFACTOR software, indicate involvement of conserved residues include 97, 99, 143, 144, 145 and 147 residues in binding site with the highest
CscoreLB (the confidence score of predicted
BS-score >1 reflects a significant local match between the predicted and template binding site. (Figure 4)
Fig: 4.Cofactor ligand binding site. involvement of conserved residues in binding site
3.7. single-scale amino acid properties assay
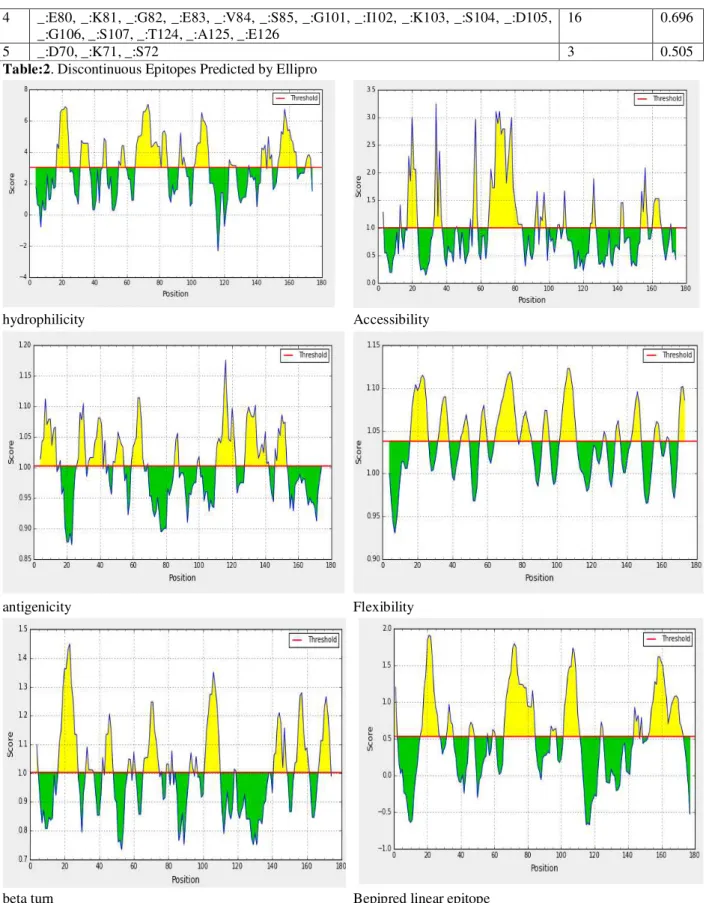
IEDB server predict several properties such as
hydrophilicity, accessibility, antigenicity,
flexibility and beta turn secondary structure in the protein sequence. Propensity scale methods assign a propensity value to each amino acid which measures the tendency of an amino acid to be part of a B-cell epitope (as compared to the background). To reduce fluctuations, the score for each target amino acid residue in a query sequence is computed as the average of the propensity values of the amino acids in a sliding window centered at the target residue. hydrophilicity,
accessibility, antigenicity, flexibility and
secondary structure properties have fundamental role in B cell epitope prediction. Relying on just one of these properties, reliable results could not be achieved. Results are shown in figure 5.
3.8. Prediction of B cell epitopes by integrated strategy
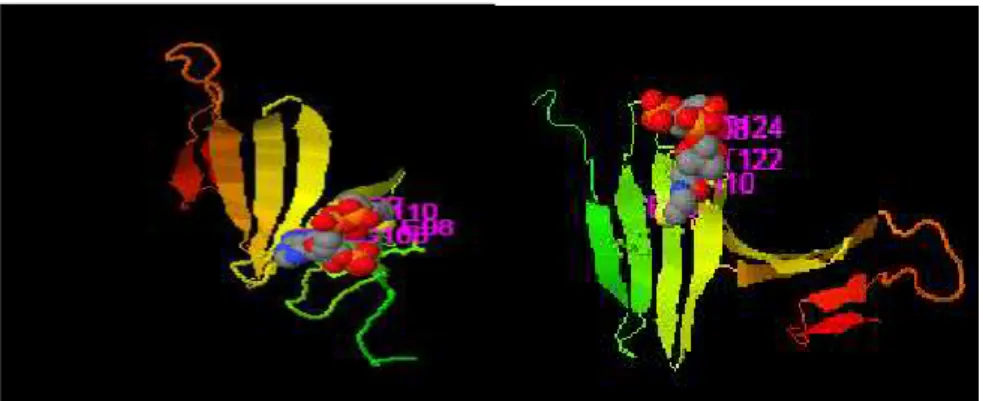
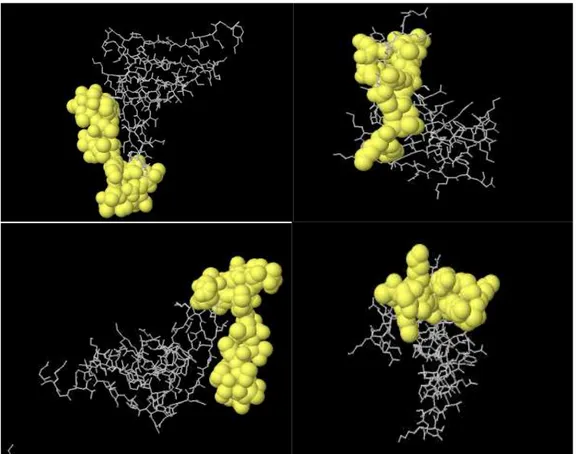
Four linear along with 11 discontinuous B cell epitopes were predicted by ElliPro software (Table 1,2). Two discontinuous and 2 linear epitopes with the highest PI (protrusion index) are shown in Figure6.
No. Start End Peptide Number of residues Score
1 55 60 KEDGKT 6 0.815
2 155 177 DTDSSAATKKTAAWNSGTSTLTI 23 0.753
3 30 37 GVKADKSK 8 0.751
4 78 84 FNEKGEV 7 0.751
5 43 47 SDDLG 5 0.733
6 1 9 DGKYDLIAT 9 0.704
7 101 107 GIKSDGS 7 0.697
8 11 25 DKLELKGTSDKNNGS 15 0.656
9 69 72 KDKS 4 0.583
Table:1. Linear Epitopes Predicted by Ellipro
no. R esiduesues Number of
residues Score
1 _:G30, _:V31, _:K32, _:A33, _:D34, _:K35, _:S36, _:K37, _:K55, _:E56, _:D57, _:G58, _:K59, _:T60
14 0.778
2 _:E134, _:G135, _:T136, _:D155, _:T156, _:D157, _:S158, _:S159, _:A160, _:A161, _:T162, _:K163, _:K164, _:T165, _:A166, _:A167, _:W168, _:N169, _:S170, _:G171, _:T172, _:S173, _:T174, _:L175, _:T176, _:I177
26 0.748
3 _:D1, _:G2, _:K3, _:Y4, _:D5, _:L6, _:I7, _:A8, _:T9, _:L13, _:E14, _:K16, _:G17, _:T18, _:S19, _:D20, _:K21, _:N22, _:N23, _:G24, _:S25, _:S43, _:D44, _:D45, _:L46, _:G47, _:K69
4 _:E80, _:K81, _:G82, _:E83, _:V84, _:S85, _:G101, _:I102, _:K103, _:S104, _:D105, _:G106, _:S107, _:T124, _:A125, _:E126
16 0.696
5 _:D70, _:K71, _:S72 3 0.505
Table:2. Discontinuous Epitopes Predicted by Ellipro
hydrophilicity Accessibility
antigenicity Flexibility
beta turn Bepipred linear epitope
Fig: 6.2 linear (up) and 2 discontinuous (down) epitopes with the highest PI score predicted by Ellipro server are shown. Epitopes mapped on 3D models using Discovery Studio Visualizer 2.5.5 software.
REFRENCES:
1. Stover, C.K., V.F. de la Cruz, T.R. Fuerst, J.E.
Burlein, I .A.Benson, L.T. Bennett, G.P. Bansal, J.F. Young, M.H. Lee, ~.F.Hatfull et al. 1991. New use of BCG for recombinant
vaccines.Nature (Lond.).351:456.
2. Aldovini, A., and R.A. Young. 1991. Humoral
and ceU-mediated immune responses to live
recombinant BCG-HIVvaccines. Nature
(Lond.).351:479.
3. Jacobs, W.R., Jr., S.B. Snapper, L. Lugnsi, and
B.R. Bloom.1990. Development of BCG as a
recombinant vaccine delivery vehicle.Cu~ Tol~
Microbiol.Immunol.155:153.
4. Jacobs, W.R., M. Tuckman, and B.R. Bloom.
1987. Introduc- tion of foreign DNA into
mycobacteria using a shuttle phasmid. Nature
(Lond.).327:532.
5. Snapper, S.B., L. Lugosi, A. Jekkel, R.E.
Melton, T. Kieser,B.R. Bloom, and W.R. Jacobs, Jr. 1988. Lysogeny and trans-formation
in mycobacteria: stable expression of foreign
genes.Proc. Natl. Acad. Sci. USA. 85:6987.
6. Newton, S.M., C,O. Jacob, and B.A. Stocker.
1989. Immune response to cholera te~in
epitope inserted in Salmonella flagellin.Science
(Wash. DC). 244:70.
7. Schorr, J., 13. Knapp, E. Hundt, H. Kupper,
and E. Amann. 1991. Surface expression of
malarial antigens in E. coli and S.typhimur/um:
induction of serum antibody response upon
oral vaccination of mice. In Vaccines
91.Modern Approaches toNew Vaccines
Including Prevention of AIDS. R.A.
Lerner,R.M. Channock, H.S. Ginsberg, and F.
Brown, editors. Cold Spring Harbor
Laboratory Press, Cold Spring Harbor,
NY.387-392.
8. Pistor, S., and G. Hobom. 1988. Expression of
viral hemag,glutinin on the surface of E. coll.
9. Bakker, D., F.G. van Zijderveld, S. van der Veen, B. Oudega,and F.K. de Graaf. 1990. K88 fimbriae as carriers of heterolo-gous
antigenic determinants. Microt~ Pathog.8:343.
10.Charbit, A., J.C. Boulain, A. Ryter, and M.
Hofuung. 1986.Probing the topology of a
bacterial membrane protein by genetic
insertion of a foreign epitope: expression at the
cell surface.EMBO (Fur. Mol. Biol. Organ.) J.
5:3029.
11.Hayashi, S., and H.C. Wu. 1990. Lipoproteins
in bacteria.J. Bioenerg.Biomembr.22:451.
12.Reiterman, A., J. Metzger, K.-H. Wiesmuller,
G. Jung, and W.G. Bessler. 1989. Lipopeptide derivatives of bacterial lipo-protein constitute potent immune adjuvants combined with or
covalently coupled to antigen or hapten. Biol.
Chem. Hoploe-Sfler. 370:343.
13.Deres, K., H. Schild, K.-H. Wiesmuller, G.
Jung, and H.-G.Ramensee. 1989. In vivo
priming of virus-specific cytotoxic T
lymphocytes with synthetic lipopeptide
vaccine. Nature (Lond.).342:561.
14.Melchers, F., V. Braun, and C. Galanos. 1975.
The lipoprotein of the outer membrane of
Escher/chia coli: a B-lymphocytemitogen. J.
ExI~ Med. 142:473.
15.Brandt, M.E., B.S. Riley, J. Rudolf, and M.V.
Norgard. 1990.Immunogenic integral
membrane proteins of Borreliaburgdor- fir/are
lipoproteins. Infect. Immu,.58:983.
16.Chamberlain, N.R., M.E. Brandt, A.L. Erwin,
J.D. Rudolf, and M.V. Norgard. 1989. Major integral membrane protein immunogens of
Treponemapallidumare proteolipids. Infect.
Immun. 57:2872.
17.Pinke, M., M. Duchene, A. Eckhardt, H.
Domdey, and B.U.yonSpecht. 1990. Protection
against experimental Pseudomonas
aeruginosainfection by recombinant P.
aeruginosalipoprotein I expressed in
Escher/chia coli. Infect. Immun. 58:2241.
18.Green, B.A., T. Quinn-Dey, and G.W.
Zlotnick. 1987. Bio-logic activities of antibody to a peptidoglycan-associated lipo-protein of
Haemophilusinfluenzaeagainst multiple
clinical isolates of H. in.fluenzaetype b. Infect.
Immu,.55:2878.
19.Hoehn, G.T., and V.L. Clark. 1992. The major
anaerobicallyinduced outer membrane protein
of Neisser/a gonorrhoeae, Pan 1, is a
lipoprotein. Infect. lmmun. 60:4704.
20.Sellwood, R., K.A. Kent, M.R. Burrows, R.J.
Lysons, and A.P. Bland. 1989. Antibodies to a
common outer envelope an-tigen of
Treponemahyodysenter/aewith antibacterial
activity. J.Gen. Microbiol. 135:2249-2257.
21.Sjostedt, A., G. Sandstrom, and A. Tarnvik.
1992. Humoral and call-mediated immunity in mice to a 17-kilodalton lipo-protein of
Fmncisellatularensis. Infect. Immun. 60:2855.
22.Bergstrom, S., V. Bundoc, and A.G. Barbour.
1989. Molecular analysis of linear plasmid encoded major surface proteins,
23. Recombinant BacilleCalmette-Guerin
Candidate Lyme Disease Vaccine OspA and OspB, of the Lyme disease spirochete
Borreliaburg-do~feri. Mol. Microbiol. 3:479.
24.Simon, M.M., U.E. Schaible, M.D. Kramer, C.
Eckerson, C.Museteanu, Hermelink, and R. Wallich. 1991.Recombinant outer surface
protein A from Borreliaburgdorferiinduces
antibodies protective against spirochetal
infection inmice..J. Infect. Dis. 164:123.
25. Fikrig, E., S.W. Barthold, F.S. Kantor, and R.
Flavell. 1990.Protection of mice against the Lyme disease agent by immunizing with
recombinant OspA. Science (Wash,
DC).250:553.
26.Fikrig, E., S.W. Barthold, N. Marcantonio, K.
Deponte, F.S. Kantor, and R. Flavell. 1992. Roles of OspA, OspB, andflagellin in protective immunity to Lyme borreliosis in
laboratory mice. Infect. Immun. 60:657.
27.Fikrig, E., S.R. Telford, S.W. Barthold, F.S.
Kantor, A.Spielman, and R.A. Flavell. 1992.
Elimination of Borreliaburgdorferifrom
vectors ticks feeding on OspA-immunized
28.Dunn, J.J., B.N. Lade, and A.G. Barbour. 1990. Outer surface protein A (OspA) from the
Lyme disease spirochete, Borreliaburgdotferi:
high level expression and purification of a
soluble recombinant form of OspA. Protein
Expression Purif1:159.
29.Erdih, L.F., M. Brandt, D.J. Warakomski, G.J.
Westrack, A.Sadziene, A.G. Barbour, andJ.P. Mays. 1993. Role of Attached Lipid in
Immunogenicity of Borreliaburgdo~feriOspA.
Infect.Immun. 61:81.
30.Bordier, C. 1981. Phase separation of integral
membrane proteins in Triton X-114 solution. J.