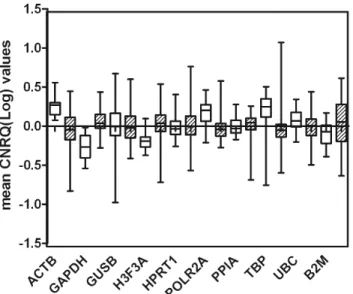
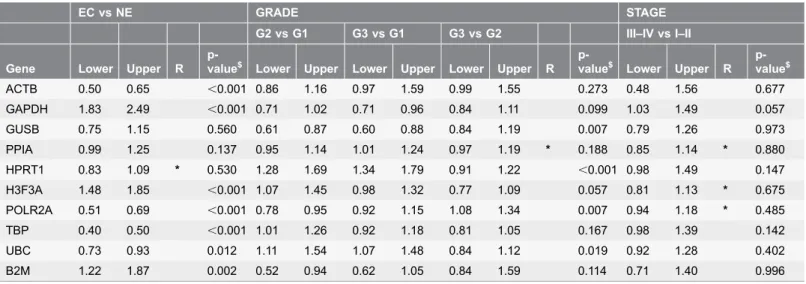
Identification of optimal reference genes for gene expression normalization in a wide cohort of endometrioid endometrial carcinoma tissues.
Texto
Imagem
Documentos relacionados
The integration of functional gene annotations in the analysis confirms the detected differences in gene expression across tissues and confirms the expression of porcine genes being
To identify the most suitable reference genes for RT-qPCR analysis of target gene expression in the hepatopancreas of crucian carp (Carassius auratus) under various conditions
This study aimed at evaluating the stability of candidate reference genes in order to identify the most appropriate genes for the normalization of the transcription in rice and red
Taking into account the results presented here, we have selected TIP41 and EF1 to be used in combination as reference genes in the forthcoming expression qPCR analysis of target
Taken all together, our results confirmed that the most stable reference genes ( GAPDH and ACTIN ) identified in current study could be used for the normalization of gene
At the first stage of the measurements results analysis, the gear wheel cast surface image was compared with the casting mould 3D-CAD model (fig.. Next, the measurements results
To confirm that alterations in the mRNA expression profiles of the metabolic genes were reflected in protein expression, we performed IHC staining for 7 metabolic genes on
Results: The analysis of gene expression data identified B2M and RPLP0 as the most stable reference genes and showed that the level of PPIA was significantly different among subjects