Identification of serum microRNA biomarkers for tuberculosis using RNA-seq.
Texto
Imagem
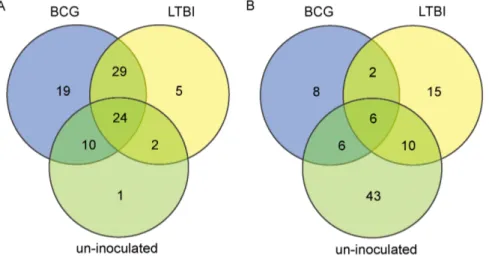
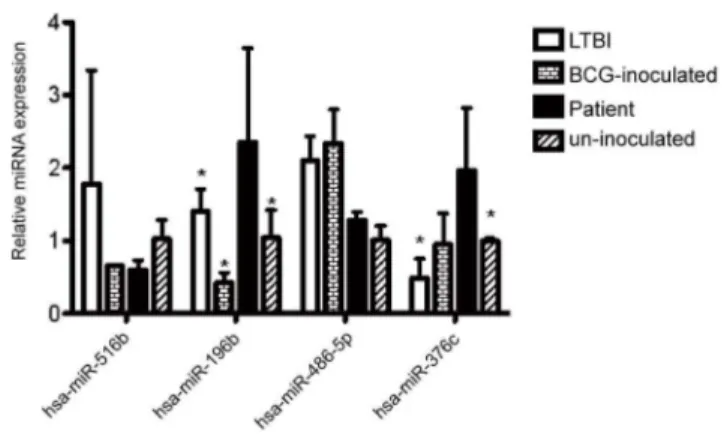
Documentos relacionados
serum and vaccine; pneumonia serum and vaccine; gangrene serum; serum for the treatment of whooping-cough; smallpox vaccine; pyogenic vaccine; streptococcus serum
RNA-Seq analysis and identification of differentially expressed genes (DEGs) - We performed RNA-Seq anal- yses to evaluate spatiotemporal gene expression profiles along
The aim of the present study was to determine the serum concentrations of 25(OH)D, serum calcium, serum phosphorus, alkaline phosphatase, and parathormone (PTH) in patients
As fold changes of upregulated microRNAs were greatly higher than that of the downregulated ones, the levels of the differentially expressed miR-3162-5p, miR- 4484, and miR-1229-5p
( C ) Western blot analysis of serum fish detected using rabbit polyclonal antibody to silver catfish IgM-like and goat anti-rabbit conjugate. In all panels the serum analyzed were:
Using miR-16 as normalization control, 3 significantly up-regulated miRNAs (miR-122, miR-222 and miR-223) and 1 significantly down-regulated miRNA (miR-21) in serum in HCC patients
The objective of the present study was to determine whether serum selenium levels are associated with the conversion of bacteriological tests in patients diagnosed with
In the present study, a proteomic approach using a label- free quantitative MS analysis identified eight differentially expressed proteins in the serum of DM1 patients compared