Analysis of Sequence Based Classifier Prediction for HIV Subtypes
Texto
Imagem
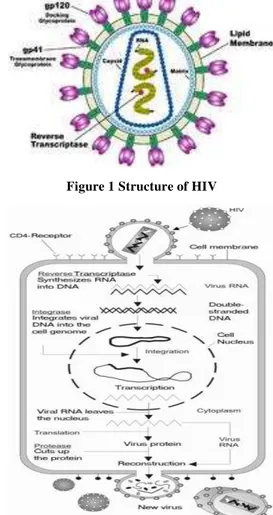

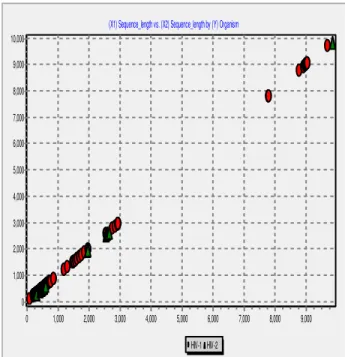
Documentos relacionados
As a decision made by the Administration Board itself: to tune and support it’s own process by a software tool to manage and monitor the requests, our thesis work so far turns out to
Based on its 16S rRNA gene sequence analysis and its morphological, physiological, and biochemical charac- teristics, strain BJ71 was identified as Cupriavidus campinensis..
Based on the results of our proposed method, fusion of classifiers on two data sets improves classification accuracy. Based on these results, proposed classifier
The first column shows the phenotypes, the second column shows the CCA association value, the third column represents the Fisher’s combined association value and the fourth
Using HIV-1 protein structures and experimental data, we evaluated the performance of individual and combined sequence-based methods in the prediction of HIV-1 intra- and
Very often, patients’ values and circumstances restrict clinicians from following guidelines word to word in all cases and they have to select one most
BLAST is a collection of sequence alignment programs from National Center for Biotechnology Information (NCBI) that uses the same heuristic approach to identify
Detection of seven major evolutionary lineages in cyanobacteria based on the 16S rRNA gene sequence analysis with new sequences of five marine Synechococcus