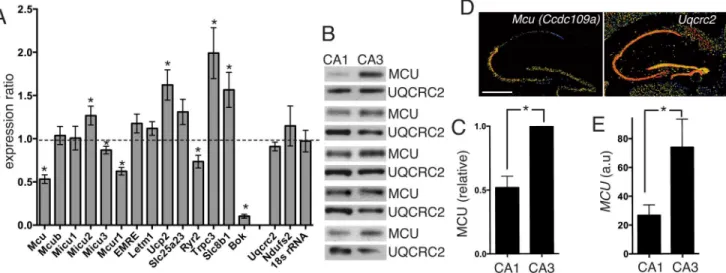
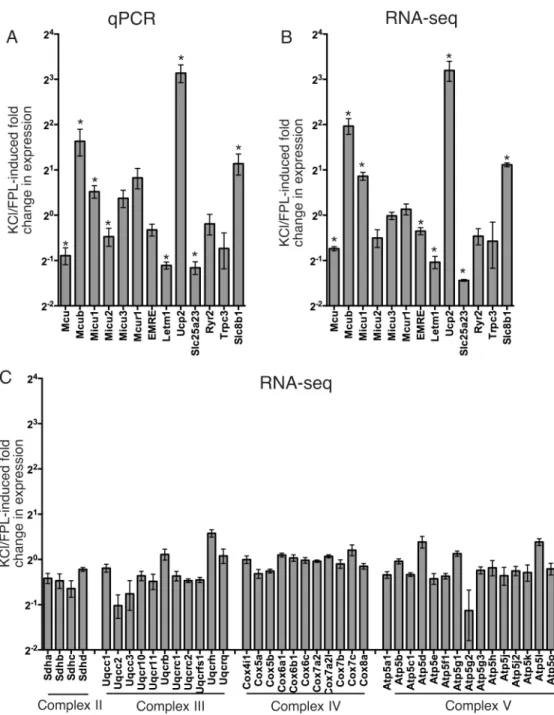
Expression of mRNA Encoding Mcu and Other Mitochondrial Calcium Regulatory Genes Depends on Cell Type, Neuronal Subtype, and Ca2+ Signaling.
Texto
Imagem


Documentos relacionados
Related enzyme activity of mitochondrial electron transport chain (METC) and creatine kinase (CK): 48 hours after surgery, the diaphragm, soleus and plantaris muscles were
To help with the interpretation of the different spectra the resonance from the phosphate buffer was used as a reference (e.g. resonances from phosphate buffer were aligned at
Results revealed an immediate switch in the expression of genes encoding alpha and beta isoforms of myosin heavy chain, and up-regulation of the cardiac isoform of alpha actin.. We
These results indicate that defective components of mitochondrial electron transport chain and oxidative phosphorylation may serve to protect cells against
At HH22 it is detected in and around the eyes (e) (Fig. mRNA expression profile of cadherin 1.. 4F) and can also be detected in the limb bud (fl, forelimb and hl, hindlimb in Fig..
We find that genes encoding members of protein complexes exhibit limited expression variation and overlap significantly with a manually derived set of dosage sensitive genes.. We
We measured the mitochondrial oxidative phosphorylation (mtOXPHOS) activities of all five complexes and determined the activity and gene expression in detail of the Complex III
In order to determine the effect of methylation change on the expression of MSAP fragments associated genes in various developmental stages during dormancy and fruit set,