Expression analysis of miRNA and target mRNAs in esophageal cancer
Texto
Imagem
Documentos relacionados
Polymorphisms of DNA repair genes XRCC1 and XPD and their associations with risk of esophageal squamous cell carcinoma in a Chinese population.. DNA repair gene XRCC1
Gene expression profiling showing differentially expressed genes between TTP -high and TTP -low expressing tumors in TCGA breast cancer ( A ), lung adenocarcinoma ( B ), lung
Purpose: To investigate the association between genetic polymorphisms of growth factor-related genes and prognosis in patients with advanced esophageal squamous cell carcinoma
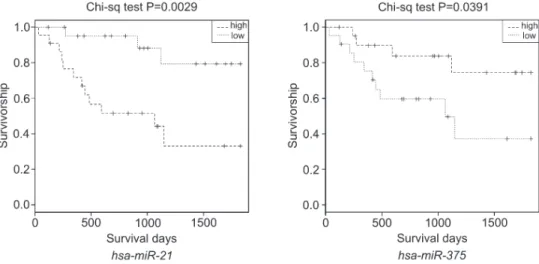
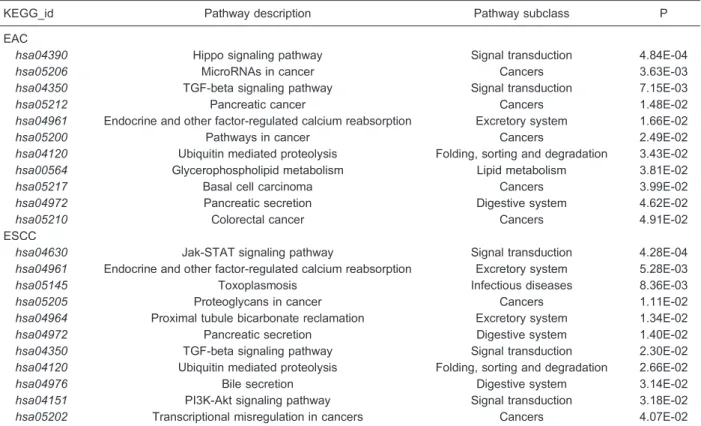
inverse regulation of expression of mRNAs and their specific miRNAs, we determined (MicroCosm Target algorithm) for each miRNA its potential target(s) and the regulatory pathways
MiRNA profiling conducted 2 weeks after Müller cell ablation identified a number of differentially expressed miRNAs whose target genes were associated with cell apoptosis
Functional analysis of significantly differentially expressed miRNAs The target gene prediction for differentially expressed miRNAs with foldchange values > 10 or < -10
The aim of this study was to examine HOTAIR expression in patients with esophageal squamous cell cancer (ESCC) and explore its clinical significance.. Methods: Differences in
Gene ontology (GO) enrichment of differentially expressed genes revealed protein folding, cell morphogenesis and cellular component morphogenesis as the top three functional terms