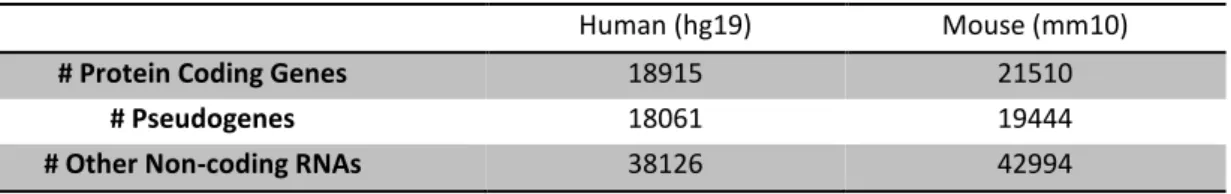
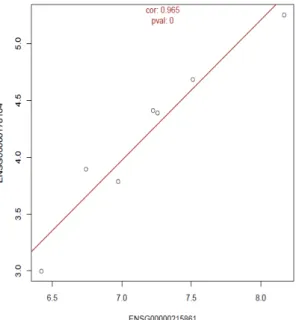
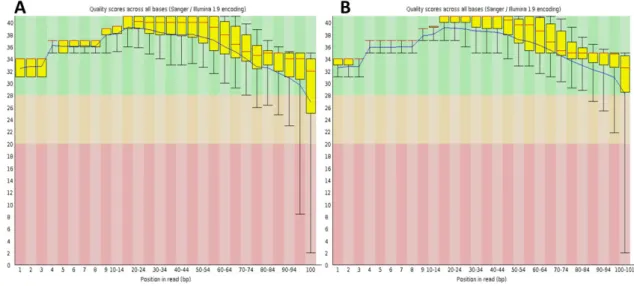
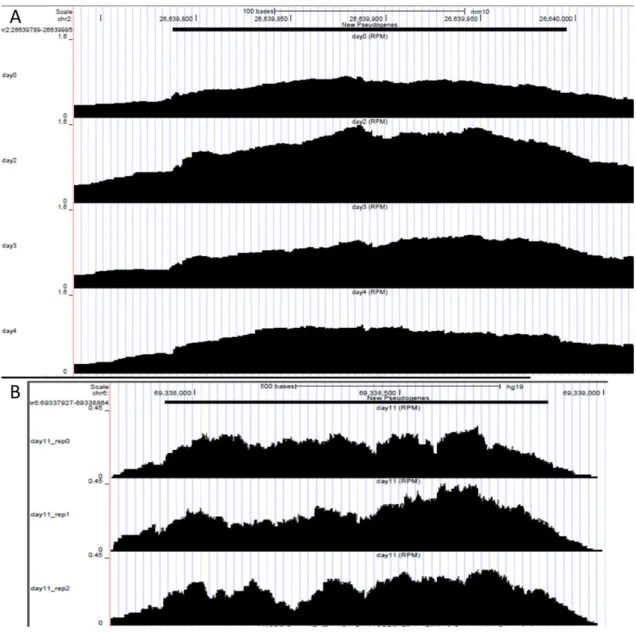
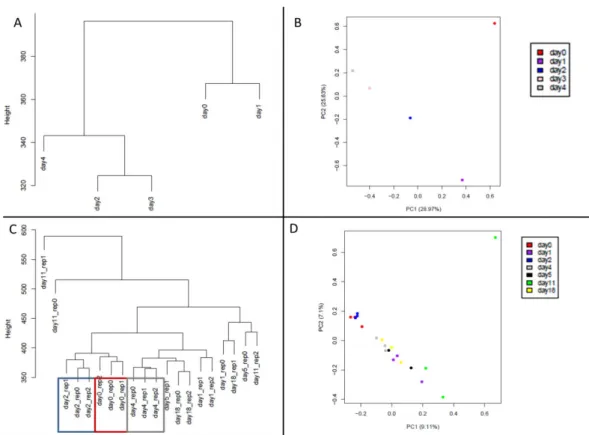
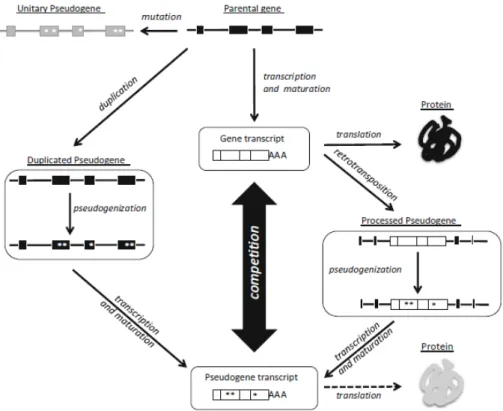
Assessing pseudogene expression during neural differentiation by RNA-Seq
Texto
Imagem
Documentos relacionados
In the present study, we identified ubiquitous and tissue-specific [Na + ] i /[K + ] i -sensitive transcriptomes by comparative analysis of differentially expressed genes in
4 ) 48 out of 90 selected differentially expressed genes were confirmed by qRT-PCR, including genes implicated in energy metabolism, neuronal differentiation, signaling,
Through DGE analysis, we obtained 1,566 differentially expressed genes in SOF/NC, and 1,099 genes in LOF/NC, where the differentially expressed genes related to lipid metabolism
We analyzed the expression of hundreds of miRNAs predicted to be expressed during myogenic differentiation using a customized microarray and identified several known and
Using data from a microarray analysis of gene expression along a time course of differentiation (Figure S1A) [22], we analyzed the linear chromosomal distribution of co-regulated
We sought to identify patterns of gene expression in the MI during pod differentiation, determine biological processes associated with pod differentiation and determination,
To facilitate an analysis of the role of BRaf kinase in brain development, postnatal neural stem cell proliferation and neuronal differentiation, we created a conditional allele
From the two different developmental stages (myce- lium and fruiting body), we identified differentially expressed genes by transcriptome analysis and found that numerous