Genome-wide analysis in human colorectal cancer cells reveals ischemia-mediated expression of motility genes via DNA hypomethylation.
Texto
Imagem

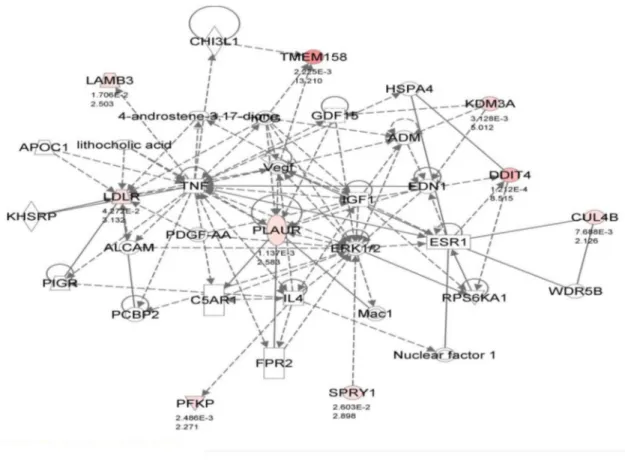

Documentos relacionados
Several upregulated genes are involved in positive reg- ulation of cell proliferation and nucleotide metabolism, whereas several downregulated genes are involved in inhibition of
A more comprehensive analysis revealed that within the greatest transcribed genes in mice infected with biofilm- released cells are genes with important functions in both innate
Specific genes that are predominantly expressed or exclusively expressed in prostate cells, prostate cancer cells, and prostate metastasis cells at the level of DNA, RNA and
We analyzed differential gene expression within the context of time-mediated gene-expression changes and show that in the context of HBV replication a number of genes and
Collectively, our analysis indi- cates that replication-dependent DNA damage may affect the expression level of a number of genes involved in cytoskeletal organization through
Our prior gene expression microarray analysis identified a number of genes that could predict metastatic behavior in thymomas [8].. Of note, these genes were not part of
In this study, we developed a computational method to identify colorectal cancer-related genes based on (i) the gene expression profiles, and (ii) the shortest path analysis
imaging showed that chitosan activated the PPAR activity in the brain and the KEGG pathway analysis showed that chitosan significantly regulated the expression of genes involved