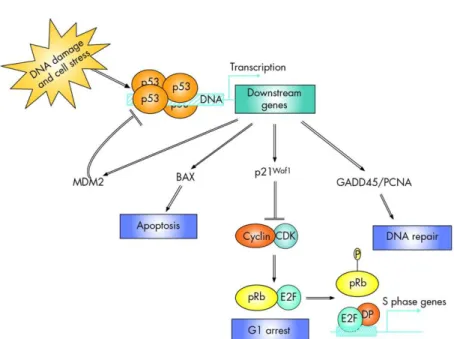
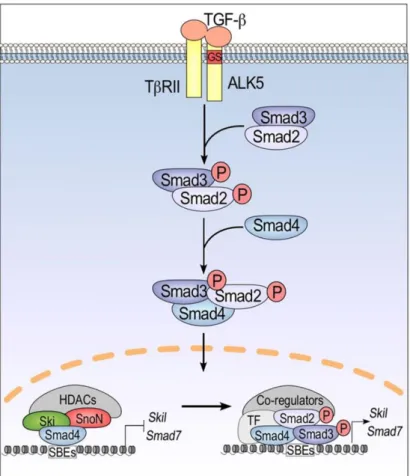
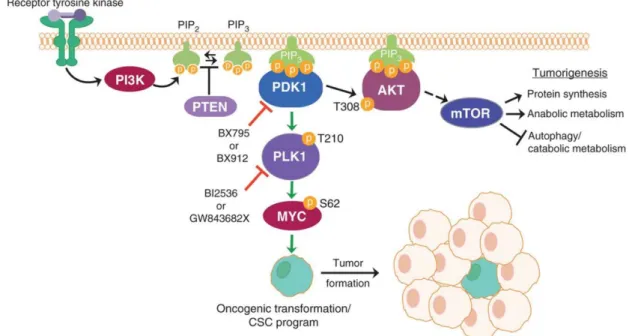
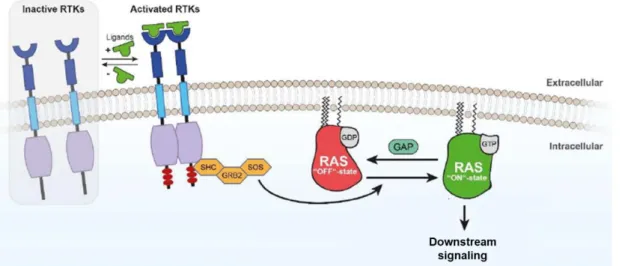
Epigenetic biomarkers as predictors of clinical outcomes in colorectal cancer
Texto
Imagem

Documentos relacionados
differential hybridization, electronic subtrac- tion (including serial analysis of gene ex- pression; SAGE), differential display reverse transcriptional-polymerase chain
Differential expression of 4 of 6 genes found to be differentially expressed by cDNA microarray analysis in the CSIII tumor versus normal tissue com- parison was validated
In the present study, we identified ubiquitous and tissue-specific [Na + ] i /[K + ] i -sensitive transcriptomes by comparative analysis of differentially expressed genes in
Through DGE analysis, we obtained 1,566 differentially expressed genes in SOF/NC, and 1,099 genes in LOF/NC, where the differentially expressed genes related to lipid metabolism
In this study, differentially expressed miRNAs in 5 pairs of gastric cancer – adjacent normal tissues (see patients ’ data in S2 Table ) were profiled using Affymetrix miRNA
The remaining two-thirds (23/36) have been reported previously as differentially expressed proteins or genes in NSCLC tumor tissue (Table 2). Novelty was determined by
The y -axes plotted in (B–F) are (B): proportion of methylated (blue) and non-methylated genes (red); (C): percentage of methylated CpG sites in methylated genes; (D): adult
Genome-wide 5mC Profiling in sALS Spinal Cord To identify loci-specific differentially methylated genes (DMGs) in sALS, postmortem spinal cord tissue (Tables 1, S1) was subjected to