Determining host metabolic limitations on viral replication via integrated modeling and experimental perturbation.
Texto
Imagem


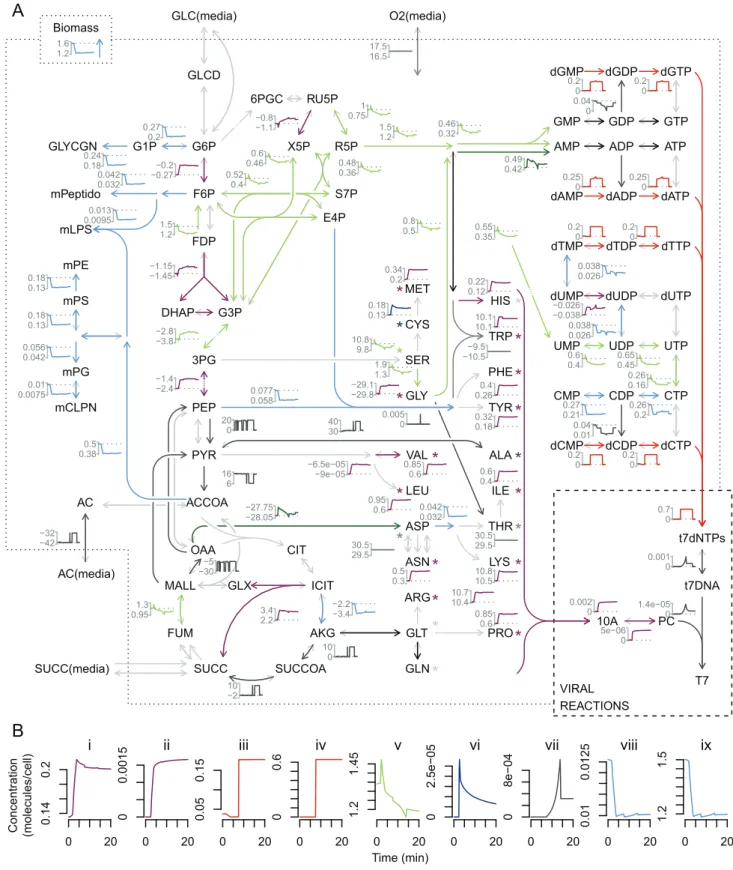

Documentos relacionados
Viral hepatitis A, B, C, D and E – systemic hepatotropic viral infections – present as acute hepatitis that, depending on the etiological agent, viral load and host conditions,
With this viral dynamics food web module (model 2), we then look at how in-host life history trade-offs mediate competition and the disease burden between virulent and
We used a mouse model of inflammatory colitis, caused by the attaching and effacing pathogen Citrobacter rodentium [14,15], to investigate dynamic changes in microbial
Therefore, to clarify the relationship between viral replication and host cytokine response against infection of the HPAIVs at an individual level, we assessed the correlation
To elucidate the in vivo involvement of PSGL-1-dependent replication and pathogenesis, and the role of amino acid polymorphism at VP1-145, we investigated viral replication,
As multiple Mimivirus capsid progeny are generated, large host cisternae that act as viral lipid reservoir are excluded from the membrane assembly zone, thus highlighting the need
Changes in the viral protein length relative to the host related proteins in the same protein Pfam family could be due to a shortening of IDOL (inter- domain linkers) and the
Human cytomegalovirus (HCMV) and rhesus cytomegalovirus (RhCMV) express a viral interleukin-10 (cmvIL-10 and rhcmvIL-10, respectively) with comparable immune modulating properties
