Hospitalization Risk Due to Respiratory Illness
Associated with Genetic Variation at IFITM3 in
Patients with Influenza A(H1N1)pdm09
Infection: A Case-Control Study
Article in PLoS ONE · June 2016
Impact Factor: 3.23 · DOI: 10.1371/journal.pone.0158181
READS
13
7 authors, including:
Pedro Pechirra
National Institute of Health Dr. Ricardo Jorge
30PUBLICATIONS 122CITATIONS
SEE PROFILE
Raquel Guiomar
National Institute of Health Dr. Ricardo Jorge
34PUBLICATIONS 714CITATIONS
SEE PROFILE
Carlos Matias Dias New University of Lisbon
104PUBLICATIONS 585CITATIONS
SEE PROFILE
Marta Barreto
National Institute of Health Dr. Ricardo Jorge
27PUBLICATIONS 298CITATIONS
RESEARCH ARTICLE
Hospitalization Risk Due to Respiratory Illness
Associated with Genetic Variation at
IFITM3
in Patients with Influenza A(H1N1)pdm09
Infection: A Case-Control Study
Vânia Gaio1, Baltazar Nunes1, Pedro Pechirra2, Patrícia Conde2, Raquel Guiomar2, Carlos Matias Dias1, Marta Barreto1*
1Departamento de Epidemiologia, Instituto Nacional de Saúde Doutor Ricardo Jorge, 1649–016 Lisboa, Portugal,2Departamento de Doenças Infeciosas, Instituto Nacional de Saúde Doutor Ricardo Jorge, 1649– 016 Lisboa, Portugal
*marta.barreto@insa.min-saude.pt
Abstract
Background
Recent studies suggest an association between the Interferon Inducible Transmembrane 3 (IFITM3) rs12252 variant and the course of influenza infection. However, it is not clear whether the reported association relates to influenza infection severity. The aim of this study was to estimate the hospitalization risk associated with this variant in Influenza Like Ill-ness (ILI) patients during the H1N1 pandemic influenza.
Methods
A case-control genetic association study was performed, using nasopharyngeal/oropharyn-geal swabs collected during the H1N1 pandemic influenza. Laboratory diagnosis of influenza infection was performed by RT-PCR, theIFITM3rs12252 was genotyped by RFLP and tested for association with hospitalization. Conditional logistic regression was performed to calculate the confounder-adjusted odds ratio of hospitalization associated withIFITM3rs12252.
Results
We selected 312 ILI cases and 624 matched non-hospitalized controls. Within ILI Influenza A(H1N1)pdm09 positive patients, no statistical significant association was found between the variant and the hospitalization risk (Adjusted OR: 0.73 (95%CI: 0.33–1.50)). Regarding
ILI Influenza A(H1N1)pdm09 negative patients, CT/CC genotype carriers had a higher risk of being hospitalized than patients with TT genotype (Adjusted OR: 2.54 (95%CI: 1.54–4.19)).
Conclusions
TheIFITM3rs12252 variant was associated with respiratory infection hospitalization but not specifically in patients infected with Influenza A(H1N1)pdm09.
a11111
OPEN ACCESS
Citation:Gaio V, Nunes B, Pechirra P, Conde P, Guiomar R, Dias CM, et al. (2016) Hospitalization Risk Due to Respiratory Illness Associated with Genetic Variation atIFITM3in Patients with Influenza A(H1N1)pdm09 Infection: A Case-Control Study. PLoS ONE 11(6): e0158181. doi:10.1371/journal. pone.0158181
Editor:Jieru Wang, University of Pittsburgh, UNITED STATES
Received:March 14, 2016
Accepted:June 10, 2016
Published:June 28, 2016
Copyright:© 2016 Gaio et al. This is an open access article distributed under the terms of the
Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability Statement:All relevant data are within the paper and its Supporting Information files.
Funding:This study was funded by the National Health Institute Doutor Ricardo Jorge. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Introduction
Influenza is a contagious and debilitating disease of the respiratory tract that can lead to hospi-talization and even death, remaining a public health threat. In April 2009, an outbreak of influ-enza A(H1N1)pdm09 infection was detected in Mexico, with subsequent cases observed in many other countries, including Portugal. It has spread quickly throughout the world and caused at least 15000 deaths in less than a year [1]. Unlike the disease pattern observed during epidemics of seasonal influenza, many of the severe cases of influenza A(H1N1)pdm09 virus infection were observed in healthy young people, with approximately 90% of all deaths due to influenza occurring in those younger than 65 years [2]. Most people infected with influenza A (H1N1)pdm09 virus experienced mild disease, with upper respiratory illness similar to sea-sonal influenza virus infection. In contrast, gastrointestinal symptoms occurred more fre-quently in patients infected with pandemic influenza virus than in those with seasonal influenza [3]. According to the Portuguese Influenza Surveillance Program (NISP), 64% of the laboratory-confirmed cases for the influenza A(H1N1)pdm09 virus were detected in the age group of 5–14 years old. A total of 1436 hospitalizations were reported, and the estimated mor-tality rate was 1.17 per 100000 inhabitants, corresponding to 124 laboratory confirmed influ-enza deaths [4].
It is commonly accepted that the severity of influenza A(H1N1)pdm09 infection results from a complex interplay between host and pathogen factors but there are many gaps in the basic understanding of what influences illness severity mainly among people without known comorbidities. While some individuals resist infection or recover quickly, others experience severe disease associated with the infection. Intensive research has been performed on the viru-lence of the virus including its genomic variability but very little is known about the host genetic background influence in the influenza outcome, despite its crucial impact on the immune response and the course of infection [5]. Consequently, in 2009, the World Health Organization identified studies of the host genetic factors’role on susceptibility to severe influ-enza as a priority [6].
Although characteristics such as age, comorbidities, degree of pre-existing immunity, immunosupression, pregnancy and smoking influence the acquisition, progression and resolu-tion of influenza infecresolu-tions, genetic variants found in human popularesolu-tions affect the specificity of virus binding and subsequent effectiveness of the immune response [7]. Their importance in humans has already been shown for several bacterial and viral pathogens [8] and it is currently known that infectious disease lethality has a heritable component [9]. In addition, animal mod-els seem to support the biologic plausibility of a genetic susceptibility to influenza A virus, given that some mouse strains are more susceptible to influenza virus infection and severity, independently of virus subtype, suggesting common infection pathways [10].
It has been shown that an interferon inducible transmembrane 3 protein (IFITM3), which acts as a viral restriction factor that mediates cellular resistance to several viruses by blocking early stages of viral replication, profoundly alters the course of influenza virus infection in a knockout mouse model. Mice lacking a functionalIfitm3gene developed severe viral pneumo-nia when challenged with normally low pathogenic viruses, and protection was re-established with reintroduction ofIfitm3. In humans, an overrepresentation of individuals with the
IFITM3rs12252 variant (C allele) that alters a splice acceptor site was found in European patients who required hospitalization as a result of influenza infection [11].
Caucasians and Han Chinese populations, concluding that the C allele constitutes a risk factor for more severe influenza infection cases [14].
Considering the wider confidence intervals, the small samples sizes and the heterogeneity in the inclusion criteria of the previous studies, the main objective of the present study was to esti-mate the association between theIFITM3rs12252 variant (C allele) and the risk of hospitaliza-tion due to respiratory illness in Portuguese patients with influenza A(H1N1)pdm09 infechospitaliza-tion using an appropriately designed epidemiological study.
Methods
Study design and participants
A case-control genetic association study, comparing the frequency ofIFITM3rs12252 variant between hospitalized influenza-like illness (ILI) patients (cases) and non hospitalized ILI patients (controls) was performed. Biological samples from cases and controls consisted on the nasopharyngeal/oropharyngeal swabs received by the Portuguese National Influenza Reference Laboratory, at the National Health Institute Doctor Ricardo Jorge (INSA) for diagnostic pur-poses during the H1N1 pandemic, from the Portuguese Laboratory Network for the Diagnosis of Influenza Infection between September 2009 and February 2010 [15].
INSA coordinated this network that was responsible for carrying out the laboratory diagno-sis of influenza A(H1N1)pdm09 infection requested by the National Health Service during the pandemic (H1N1)2009. This network received swabs from ILI patients identified by primary healthcare units and hospitals. All information collected regarding clinical and epidemiological information, such as patients’demographic data, signs and symptoms and underlying condi-tions and laboratory results were stored in a national common database. A total of 62089 ILI cases were reported to the laboratory network during the H1N1 pandemic. From these 25985 were laboratory-confirmed influenza cases, over 99% of which were associated with the new pandemic strain A(H1N1)[15]. At INSA 16376 samples for Influenza diagnosis were received. Of those, 7780 influenza A(H1N1)pdm09 positive cases were identified.
An ILI positive patient was defined as a patient presenting signs and symptoms complying to the European ILI case definition [16], who was swabbed and tested positive for influenza A (H1N1)pdm09 using real-time polymerase chain reaction (RT-PCR) [17]. ILI negative patients were ILI patients who were swabbed and tested negative for the Influenza A(H1N1)pdm09 virus. ILI patients tested negative for Influenza A(H1N1)pdm09 virus also tested negative for the Influenza A and B virus subtypes.
All included patients were selected from the INSA cases, whose tests for Influenza diagnosis were performed at National Influenza Reference Laboratory (INSA). The following exclusion criteria were used to perform the selection: (1) patients over 65 years old; (2) immunodepressed or transplanted patients (3) patients with chronic diseases (diabetes, lung, kidney, cardiovascu-lar, liver, neurological, immunologic and oncologic diseases); (4) pregnant women; (5) notifica-tion data before the mitiganotifica-tion phase (01-09-2009); (6) patients for whom the time lapse between the symptoms onset and the sample collection was over 7 days, to avoid misclassifica-tion due to low viral load of the sample collected; (7) samples with unavailable informamisclassifica-tion about hospitalization; (8) samples with unavailable laboratorial result for A(H1N1)pdm09 virus; (9) samples stored outside of the National Health Institute Doutor Ricardo Jorge.
After the exclusion criteria application, 312 hospitalized patients were identified. These cases were stratified in two groups: hospitalized Influenza A(H1N1)pdm09 positive cases (n = 108) and hospitalized Influenza A(H1N1)pdm09 negative cases (n = 204). Since the risk of developing Influenza varies with time and to assure that the selected control group was at similar risk of developing the health outcome of interest, for each case in the group of
hospitalized cases, 2 non-hospitalized controls matched for the week of onset of symptoms were selected from the Portuguese Laboratory Network database (Fig 1). Hospitalization was used as a measure of disease severity. In this context, it was defined as a hospital admission due to ILI complications with a concomitant hospital stay for more than twenty-four hours. The study protocol was approved by the Ethics Committee of the National Health Institute Doutor Ricardo Jorge. Since this study used irreversibly anonymized samples from the NPIS, obtaining informed consent from the participants was impossible and therefore the study was authorized by the Ethics Committee of the National Health Institute Doutor Ricardo Jorge without informed consent by the participants.
Laboratory diagnosis
For laboratory diagnosis and further virological characterization, the nasopharyngeal/oropha-ryngeal swabs were collected into a suitable transport medium for preservation of virus, molec-ular diagnostic and virus culture. Real-time reverse transcriptase PCR methodologies and platforms were used for laboratory diagnosis of influenza infection [17]. Additional diagnoses, viral isolation, antigenic and genetic characterization of viral isolates were performed according to the procedures previously described [15]. During the 2009/10 influenza season, the labora-tory network just carry out the laboralabora-tory diagnosis of influenza A(H1N1)pdm09 infection requested by the National Health Service and the samples were only tested for the influenza A (H1N1)pdm09, influenza A(H1), influenza A(H3) and influenza B virus.
Genotyping
Genomic DNA was extracted from the nasopharyngeal/oropharyngeal swabs using an auto-matic system (MagNA Pure LC—Roche) with the MagNA Pure LC Total Nucleic Acid Isola-tion Kit. The rs12252 SNP in theIFITM3gene was genotyped by restriction fragment length polymorphism (RFLP) using the following forward and reverse primers:TGAGGGTTATGGGA GATGGGGT(F) andGGAGAGGAGATGGTGAGGGGA(R) in standard PCR conditions. The
restriction enzymeMscIwas used to cut the PCR product in the presence of the wild type T allele and the originated fragments were visualized on a 2% agarose gel.
Fig 1. Schematic representation of the sampling process.
Statistical analysis
Statistical analysis was performed using the R program [18].P-values<0.05 were considered to
be statistically significant. The T-test and the Wilcoxon test were used to access differences of quantitative variables according to their adherence to the normal distribution. Proportions were compared using the Pearson's chi-squared test.
Regarding age and sex, the National Hospitalization Discharge (NHD) database was used to assess if the sample of influenza positive hospitalized cases analyzed in the present study was a representative of the total influenza (International Codifications of Diseases code 487.0) hospi-talized patients in the NHS, during the same period of time and excluding patients over 65 years, with chronic diseases and pregnant women. Allelic and genotypes frequencies were obtained by direct count and were tested for the Hardy-Weinberg Equilibrium, using the Hardy Weinberg R package [19]. To perform the case-control association analysis, hospitalized patients were considered the cases and non-hospitalized patients were considered the controls. The analysis was stratified by ILI A(H1N1)pdm09 positive and negative patients. To estimate the AdjustedOdds-Ratio, we used Conditional Logistic Regression for matched Pairs Data including the potential confounding effect of age and gender variables.
Results
From the 936 patients selected, 909 had sufficient available frozen swabs for DNA extraction. We successfully genotyped 792 samples: 268 from ILI A(H1N1)pdm09 positives patients (84 hospitalized cases and 184 non-hospitalized controls) and 524 from ILI A(H1N1)pdm09 nega-tives patients (173 hospitalized cases and 351 non-hospitalized controls) corresponding to an overall success rate of 87% (95% CI: 84.8%–89.2%) that was similar in all the 4 groups of patients (p= 0.998) (S1 Table).
No significant differences were found regarding the age (p= 0.382) and sex (p= 0.468) dis-tributions between cases and controls among ILI A(H1N1)pdm09 positive patients and ILI A (H1N1)pdm09 negative patients (Table 1). No significant differences were found between the analysed influenza positive hospitalized cases and influenza diagnosed hospitalized patients present in the NHD database, regarding age (p= 0.320) and sex (p= 0.133) (S2 Table).
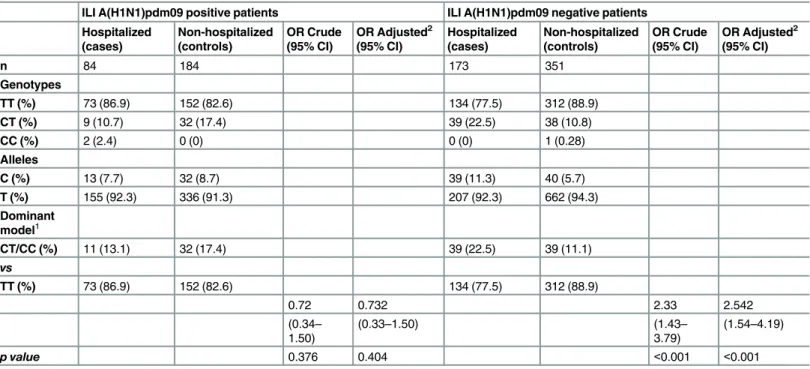
ConcerningIFITM3rs12252 variant characterization (Table 2), 3 patients carrying the CC genotype were detected (2 were ILI A(H1N1)pdm09 positive hospitalized patients and 1 was a ILI A(H1N1)pdm09 negative non-hospitalized patient).
Regarding A(H1N1)pdm09 infected patients, 10.7% (95% CI: 4.1–17.3) of the hospitalized cases and 17.4% (95% CI:11.9–22.9) of the non-hospitalized controls had the CT genotype.
Table 1. Patients characterization regarding age and gender variables.
ILI A(H1N1)pdm09 positive patients ILI A(H1N1)pdm09 negative patients
Hospitalized (cases) Non-hospitalized (controls) p Hospitalized (cases) Non-hospitalized (controls) p
Age (years)
Mean±sd 16.6±17.6 14.0±12.4 0.8151 15.7±20.2 13.5±16.1 0.3821
Median (range) 10 (0–60) 9 (0–54) 4 (0–64) 6 (0–62)
Gender
% of women 40.5 46.2 0.3822 43.9 47.3 0.4682
(95% CI) (30.0–51.0) (39.0–53.4) (36.5–51.3) (42.1–52.5)
1p-values
were obtained by the Wilcoxon test. 2p-values
were obtained by the Pearson's chi-squared test. (CI, Confidence interval;p, p-value; sd, standard deviation). doi:10.1371/journal.pone.0158181.t001
The hospitalized A(H1N1)pdm09 infected cases had a C allele frequency of 7.7% (95% CI: 3.6–
11.8) while the non-hospitalized controls had a C allele frequency of 8.7% (95% CI: 5.8–11.6). In the A(H1N1)pdm09 negative patients, 22.5% (95% CI:16.3–28.8) of the hospitalized cases and 10.8% (95% CI: 7.6–14.1) of the non-hospitalized controls carried the CT genotype. The hospitalized cases have a C allele frequency of 11.3% (95% CI: 8.0–14.6) while the non-hospi-talized controls presented a C allele frequency of 5.7% (95% CI: 2.2–9.2) (Table 2).
To assess if there is a higher risk of hospitalization for patients who carry the C allele both in the presence and absence of the A(H1N1)pdm09 virus infection, we performed a stratified analysis assuming a dominant model, as presented inTable 2. In the presence of A(H1N1) pdm09 virus infection, we observed that there is not a higher risk of being hospitalized for patients with the CT/CC genotype compared to the TT genotype (Adjusted Odds Ratio: 0.73 (95% CI: 0.33–1.50). However, when we use the same approach to assess if there is a higher risk of being hospitalized for the C allele carriers in patients negative for A(H1N1)pdm09 virus infection we found that the risk of being hospitalized among the CT/CC genotype carriers is significantly higher than the risk of being hospitalized in the TT genotype carriers (Adjusted Odds Ratio: 2.54 (95% CI: 1.54–4.19)).
Discussion
In order to measure the association between theIFITM3rs12252 variant and influenza A(H1N1) pdm09 severity infection among ILI patients without reported comorbilities, we have designed a specific case-control study, in the H1N1 pandemic scenario. Our results show that theIFITM3
rs12252 variant was associated with respiratory disease hospitalizations but not in the ILI patients with a laboratory confirmed Influenza A(H1N1)pdm09 infection. On the other hand,
Table 2. IFITM3rs12252 genotypic and allelic frequencies among the four groups of patients and its association with hospitalization, assuming a dominant model.
ILI A(H1N1)pdm09 positive patients ILI A(H1N1)pdm09 negative patients
Hospitalized (cases) Non-hospitalized (controls) OR Crude (95% CI)
OR Adjusted2
(95% CI) Hospitalized (cases) Non-hospitalized (controls) OR Crude (95% CI)
OR Adjusted2
(95% CI)
n 84 184 173 351
Genotypes
TT (%) 73 (86.9) 152 (82.6) 134 (77.5) 312 (88.9)
CT (%) 9 (10.7) 32 (17.4) 39 (22.5) 38 (10.8)
CC (%) 2 (2.4) 0 (0) 0 (0) 1 (0.28)
Alleles
C (%) 13 (7.7) 32 (8.7) 39 (11.3) 40 (5.7)
T (%) 155 (92.3) 336 (91.3) 207 (92.3) 662 (94.3)
Dominant model1
CT/CC (%) 11 (13.1) 32 (17.4) 39 (22.5) 39 (11.1)
vs
TT (%) 73 (86.9) 152 (82.6) 134 (77.5) 312 (88.9)
0.72 0.732 2.33 2.542
(0.34– 1.50)
(0.33–1.50) (1.43–
3.79)
(1.54–4.19)
p value 0.376 0.404 <0.001 <0.001
1Other genetic models were not considered due the low frequency of CC genotype and the existence of zero patients with CC genotypes in 2 groups. 2
Adjustment for age and gender by logistic regression.
we observed that the risk of being hospitalized for the CT/CC genotype was 2.54 fold when com-pared to the TT genotype in ILI patients negative for influenza A(H1N1)pdm09 infection.
Given these results, we hypothesize that this variant is involved in the ILI symptoms severity associated with other respiratory infections. During the H1N1 pandemic influenza season, the Portuguese Laboratory Network just carried out the influenza virus detection (influenza A (H1N1)pdm09, influenza A(H1) seasonal, influenza A(H3) and influenza B virus), therefore we must consider the hypothesis of the presence of other infectious agents associated with respiratory disease (adenovirus, coronavirus, rhinovirus, respiratory syncytial virus, metapneu-movirus, parainfluenza virus,Haemophilus influenzae,Mycoplasma pneumoniae,Streptococcus pneumoniaeand other less frequent respiratory agents) in the Influenza negative samples that could explain our results. In addition, some studies show thatIFITM3is also important in the mediation of other virus infections such as West Nile Virus and Dengue Virus [20] and this hypothesis could justify the higher percentage of C allele carriers in the ILI hospitalized Influ-enza negative cases. The future detection of other respiratory virus in the analyzed samples would be important to clarify our results.
When comparing our work with the study previously published by Everitel al(2012) [11] on this SNP regarding European patients, we found a similar percentage of C allele carriers among the influenza hospitalized positive cases (13.2% versus 13.1%). However, the compari-son group were not ILI non hospitalized Influenza A(H1N1)pdm09 positive patients, meaning that they did not test the hypothesis that theIFITM3rs12252 variant was involved in the sever-ity of the Influenza infection. More specifically, when comparing the general population with-out infection (European 1000 genomes) with the ILI hospitalized influenza positive patients they are evaluating both infection susceptibility and severity without the ability to distinguish each effect.
The percentage of the C allele carriers in the ILI non-hospitalized Influenza A(H1N1) pdm09 negative controls is the closest to the general population (European 1000 genomes), as expected (Fig 2).
Our results must be interpreted taking into account some limitations that are specific of the study design. The case definition (“hospitalization”) used to measure disease severity might not be considered as the best measure in a pandemic scenario given that less severe cases were likely to be hospitalized due to the initial pandemic alert, reducing the effect ofIFITM3
rs12252 on the risk of severity (hospitalization). However, we have reduced this bias by exclud-ing patients with disease onset reported in the contention phase when health authorities had the indication to hospitalise all influenza A(H1N1)pdm09 cases. In addition, the hospitaliza-tion criteria that were used in the different health units might not be uniform and could be dif-ferent from influenza A(H1N1)pdm09 positive and negative cases given the pandemic alert and the indications of the health authorities This differential criterion for hospitalization could explain the presence of theIFITM3rs12252 association in the negative influenza patients and the absence in the positive influenza patients. The extent of this bias could not be evaluated with the available data. On the other hand, we only performed the confounding adjustment for age and sex variables in the logistic regression analysis, but the reduction of possible confound-ing bias were also achieved by the exclusion criteria used in the patients selection, excludconfound-ing patients with known risk factors for influenza infection complication. Regarding the possible selection bias, we were only able to evaluate the representativeness of the influenza hospitalized patients with less 65 years of age without known co morbidities, the patients recorded in the NHD during H1N1 pandemic were not statistical different from the ones included in the study regarding age and sex variables.
Due the low frequency of theIFITM3rs12252 observed in the Portuguese population and also in the European population (3%), a large number of cases would be needed to better define
this association in our population. In the future, it will be important to perform an integrative approach to clarify why some healthy individuals resist infection or recover quickly, others experience severe disease associated with the influenza infection. It could be done by consider-ing not only the virus-host genome interactions but also immunity, vaccination, weather con-ditions and other environmental factors that could help to clarify the present results.
Moreover, early screening for the host genetic background could be a promising strategy to identify potential therapeutic targets and might help to evaluate and predict the severity of dis-ease thereby providing critical information for decision making during treatment [21].
In conclusion, this study showed that theIFITM3rs12252 variant was associated with the hospitalization risk in the ILI hospitalized Influenza A(H1N1)pdm09 negative patients. How-ever, the same association was not found in the ILI hospitalized Influenza A(H1N1)pdm09 positive cases and we cannot confirm that this variant is involved in the ILI symptoms severity associated with Influenza A(H1N1)pdm09 infection and the absence of the effect in this group could be due to bias associated with hospitalization criteria during the pandemic.
Supporting Information
S1 Table. Genotyping success rates in each ILI patients group.
(DOCX)
S2 Table. Age and sex comparison between the ILI hospitalized Influenza A(H1N1)pdm09 positive cases and the DRG patients.
(DOCX)
Fig 2. Percentage of C allele carriers (CC or CT genotypes) in the different samples.General population data are from the European 1000 genomes (Data from [11] were also included).
Acknowledgments
We acknowledge all the Colleagues from Infectious Disease Department from National Insti-tute of Health Doctor Ricardo Jorge (INSA) that supported influenza A(H1N1)pdm09 diagno-sis during 2009 Pandemia and 2009/2010 influenza season, to address the excess samples sent to National Influenza Reference Laboratory at INSA.
We especially acknowledge all management support, during 2009 Pandemia, to Doctor Jaime Nina head of Infectious Diseases Department and Doctor Cristina Furtado head of Ref-erence and Epidemiological Surveillance Unit, during 2009 and 2010.
Author Contributions
Conceived and designed the experiments: VG BN CMD MB. Performed the experiments: VG PP PC RG. Analyzed the data: VG BN MB. Contributed reagents/materials/analysis tools: MB RG. Wrote the paper: VG BN PP PC RG CMD MB.
References
1. World Health Organization (2010). Pandemic (H1N1) 2009—update 112. Available at:http://www.who.
int/csr/don/2010_08_06/en/index.html. Accessed 26 February 2016.
2. Bautista E, Chotpitayasunondh T, Gao Z, Harper SA, Shaw M, Uyeki TM, et al. (2010) Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection.
3. Team NS-OIAVI (2009) Emergence of a Novel Swine-Origin Influenza A (H1N1) Virus in Humans. New England Journal of Medicine 360: 2605–2615. doi:10.1056/NEJMoa0903810PMID:19423869
4. Froes F, Diniz A, Falcão I, Nunes B, Catarino J (2014) Final report on the mortality from flu pandemic (H1N1) 2009 in Portugal (April 2009–August 2010). Revista Portuguesa de Saúde Pública 32: 55–60.
5. Arcanjo A, Mazzocco G, de Oliveira S, Plewczynski D, Radomski J (2014) Role of the host genetic vari-ability in the influenza A virus susceptibility. Acta Biochimica Polonica 61: 403–419. PMID:25184407
6. König R, Stertz S, Zhou Y, Inoue A, Hoffmann H-H, Bhattacharyya S, et al. (2010) Human host factors required for influenza virus replication. Nature 463: 813–817. doi:10.1038/nature08699PMID: 20027183
7. Nicholson KG, Wood JM, Zambon M (2003) Influenza. Lancet 362: 1733–1745. PMID:14643124
8. Hill AV (2006) Aspects of genetic susceptibility to human infectious diseases. Annu Rev Genet 40: 469–486. PMID:17094741
9. Srivastava B, Błażejewska P, Heßmann M, Bruder D, Geffers R, Mauel S, et al. (2009) Host genetic background strongly influences the response to influenza a virus infections. PloS one 4: e4857. doi:
10.1371/journal.pone.0004857PMID:19293935
10. Boon AC, deBeauchamp J, Hollmann A, Luke J, Kotb M, Rowe S, et al. (2009) Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. Journal of virol-ogy 83: 10417–10426. doi:10.1128/JVI.00514-09PMID:19706712
11. Everitt AR, Clare S, Pertel T, John SP, Wash RS, Smith SE, et al. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484: 519–523. doi:10.1038/nature10921PMID: 22446628
12. Zhang Y-H, Zhao Y, Li N, Peng Y-C, Giannoulatou E, Jin RH, et al. (2013) Interferon-induced trans-membrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individu-als. Nature communications 4: 1418. doi:10.1038/ncomms2433PMID:23361009
13. Mills TC, Rautanen A, Elliott KS, Parks T, Naranbhai V, Ieven MM, et al. (2014) IFITM3 and susceptibil-ity to respiratory viral infections in the communsusceptibil-ity. Journal of Infectious Diseases 209: 1028–1031. doi:
10.1093/infdis/jit468PMID:23997235
14. Yang X, Tan B, Zhou X, Xue J, Zhang X, Wang P, et al. (2015) Interferon-Inducible Transmembrane Protein 3 Genetic Variant rs12252 and Influenza Susceptibility and Severity: A Meta-Analysis.
15. Portuguese Laboratory Network for the Diagnosis of Influenza Infection. Contribution of the Portuguese Laboratory Network for the Diagnosis of Influenza A(H1N1)pdm09 Infection during the 2009/10 and 2010/11 influenza seasons. Euro Surveill. 2012; 17(27):pii = 20211. Available at:http://www.
eurosurveillance.org/ViewArticle.aspx?ArticleId=20211. Accessed 10 December 2015.
16. European Centre for Disease Prevention and Control (ECDC). Overview of Surveillance Influenza 2009/2010 in EU/EEA.Technical document. Stockholm, 2009. Available at:http://www.ecdc.europa.
eu/en/publications/Publications/0909_TED_Overview_of_Surveillance_of_Influenza_2009-2010_in_
EU-EEA.pdf. Accessed 26 February 2016.
17. CDC protocol of realtime RTPCR for influenza A(H1N1). Available at:http://www.who.int/csr/
resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf.
Accessed 26 February 2016.
18. R Development Core Team (2011) R: a language and environment for statistical computing. R Founda-tion for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. Available at:http://www.R-project. org. Accessed 10 Dcember 2015.
19. Graffelman J: HardyWeinberg: Graphical tests for Hardy-Weinberg equilibrium. R package version 1.4.1. 2012. Available at:http://CRAN.R-project.org/package=HardyWeinberg. Accessed 10 Decem-ber 2016.
20. Brass AL, Huang I-C, Benita Y, John SP, Krishnan MN, Feeley EM, et al. (2009) The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139: 1243–1254. doi:10.1016/j.cell.2009.12.017PMID:20064371
21. Lander ES (2011) Initial impact of the sequencing of the human genome. Nature 470: 187–197. doi:


![Fig 2. Percentage of C allele carriers (CC or CT genotypes) in the different samples. General population data are from the European 1000 genomes (Data from [11] were also included).](https://thumb-eu.123doks.com/thumbv2/123dok_br/16720203.745000/9.918.73.870.120.547/percentage-carriers-genotypes-different-general-population-european-included.webp)