Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection.
Texto
Imagem

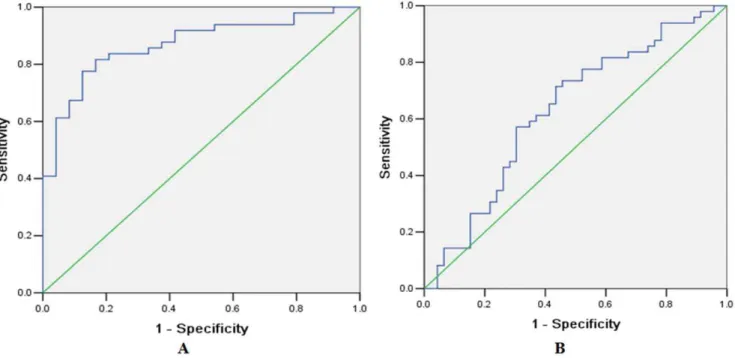
Documentos relacionados
O aumento na transcrição e consequente fosforilação de AMPKαβ, isoforma predominante no músculo esquelético, ativa processos catabólicos tais como a captação de glicose
To test whether mammary epithelial cells also release the identified signature miRNAs miR-451, miR-1246, miR-720, and miR-16 in the human body, we focused on fluids that are
Our microarray- based analysis of miRNA expression in mice cerebral cortex after TBI revealed that some miRNAs such as miR-21, miR-92a, miR- 874, miR-138, let-7c, and miR-124 could
Although there was no difference in saliva samples between groups, according to tissue and serum samples miR-21 and 30e may have an important role; since they were down-regulated
We compared the abundance of miR-29 family in urinary supernatant, as shown in Fig 1., miR-29a and miR-29c levels are significantly higher than miR-29b in urinary supernatant of type
The serum levels of miR-3200, miR-21, and miR-205 at any point during chemotherapy treatment were not predictive for DFS; however, high levels of miR-451 at the time of diagnosis
The results showed that the serum relative levels of only 3 miRNAs (miR-34a-5p, miR-221-3p and let-7d-3p) in MDD patients were obviously higher than these in control patients, and
Fig. A single nucleotide difference in seed region of miRNAs is associated with a major change in regulated target genes. A) miR-25 and miR-32, two miRNAs with identical seed