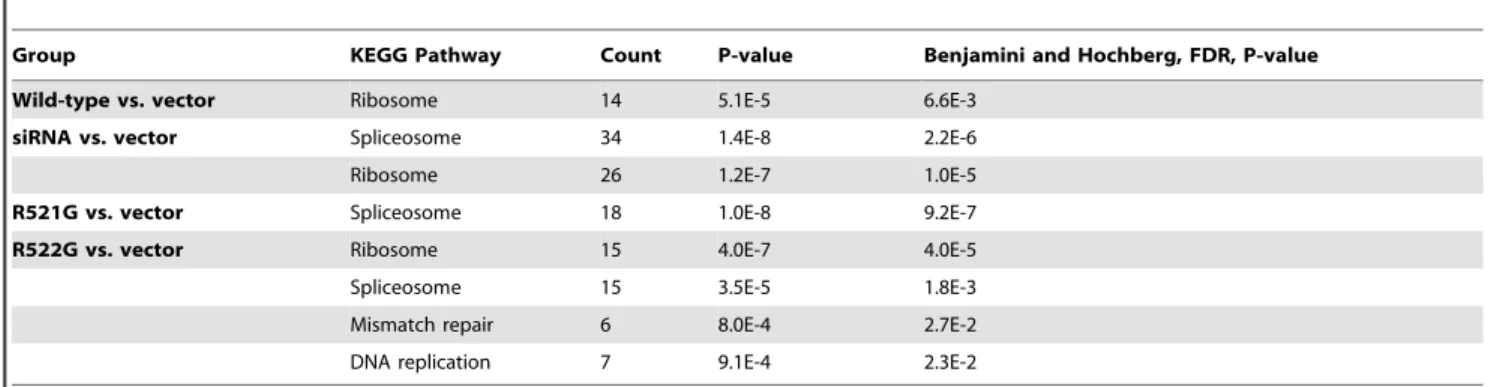
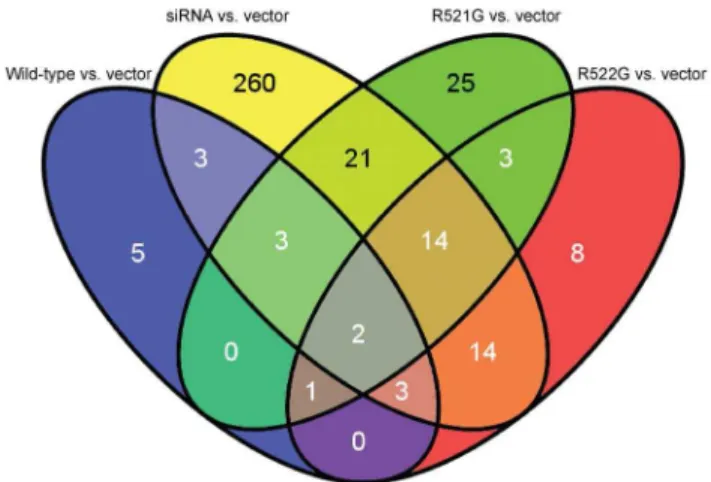
Characterization of FUS mutations in amyotrophic lateral sclerosis using RNA-Seq.
Texto
Imagem


Documentos relacionados
The broad objective of the study is to analyze male and female access to land for cassava production in Abia state and specifically to describe the
As shown in the results, sugarcane bagasse was able to activate the expression of a different set of genes that were differentially expressed compared with the control, and
Analysis of the number of genes differentially expressed in response to radiation at the two different doses was consistent with the idea that gene expression levels are dependent
Objective: To describe the prevalence and severity of periodic limb movements during sleep in amyotrophic lateral sclerosis patients and to explore this fact as a predictor
In summary, RNA-seq allowed for the transcriptomic analysis of differentially expressed genes in host cell in vitro , during CaHV-1 infection. , PTGS2 ) expression from
As a result of gene expression analysis based on whole blood RNA, the number of expressed genes was less in fetal stage and infancy period but increased with age, presenting a
Mutations in the angiogenic factor, angiogenin ( ANG ), have been identified in patients with both familial and sporadic amyotrophic lateral sclerosis (ALS) and are thought to have
In order to determine the effect of methylation change on the expression of MSAP fragments associated genes in various developmental stages during dormancy and fruit set,