RNA-Seq gene profiling--a systematic empirical comparison.
Texto
Imagem
Documentos relacionados
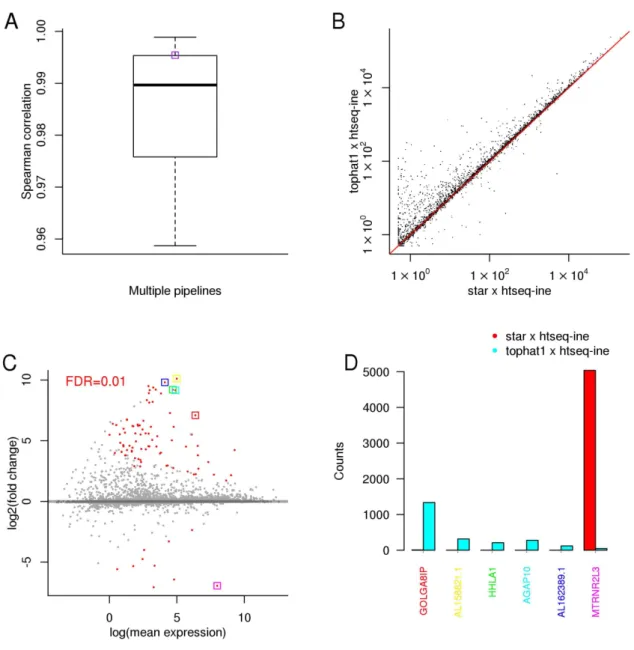
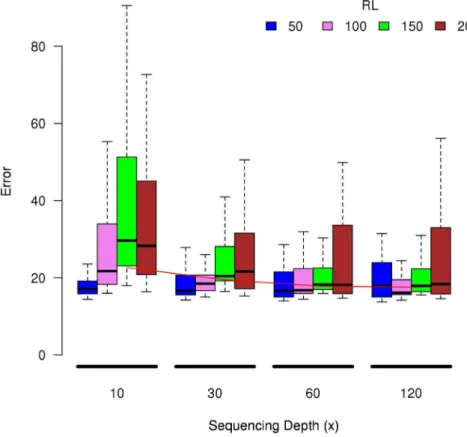
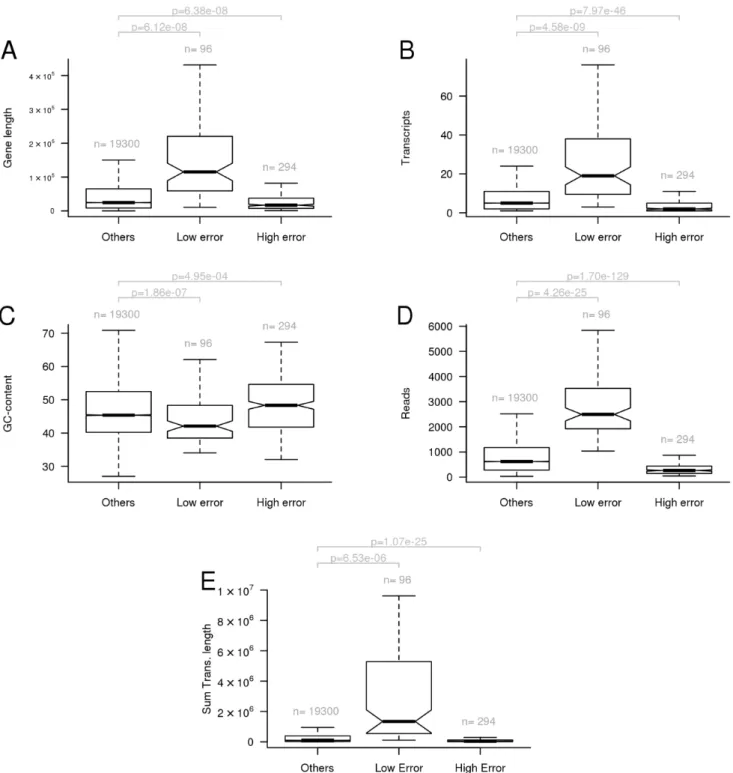
RNA-Seq analysis and identification of differentially expressed genes (DEGs) - We performed RNA-Seq anal- yses to evaluate spatiotemporal gene expression profiles along
Relative gene expression for all six genes measured by RT-qPCR was compared with RNA-Seq data on days 2 and 5 for the July experiment.. In the first comparison relative expression
Based on RNA-seq analysis, we recently found that the NCAPG gene and its neighboring gene, LCORL , are both differentially expressed in longissimus muscle of fetal and adult
In the current study, we performed chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) to evaluate the effects of sorafenib on the genome-wide profiling
To identify additional accumulated mRNAs in the nucleus of neurons expressing fragile X premuta- tion rCGG repeats, we performed gene expression profiling using total RNAs and RNAs
Methods: Gene expression analysis was performed on five different mouse embryonic fibroblasts (MEFs) samples (i) the Mta1 wild type, (ii) Mta1 knock out (iii) Mta1 knock out in
For the unsupervised analysis and the supervised analyses (except that centered on the mitotic index), samples were divided in a learning set including our series pooled with West
Our systematic analysis utilized a combination of comparative genomic scanning, functional pathway analysis and gene expression profiling to uncover previously unidentified