Alla andra mitokondriella proteiner kodas av gener i kärngenomet, inklusive alla enzymer som behövs för att replikera, transkribera och översätta mtDNA. Organellen innehåller också faktorer som är nödvändiga för att upprätthålla integriteten hos mtDNA-molekylen och säkerställa korrekt topologi. I denna avhandling använder vi en kombination av in vitro biokemi och cellbiologi för att förstå de underliggande molekylära mekanismerna i mitokondriell sjukdom.
BIOLOGY OF MITOCHONDRIA
- Origin of mitochondria
- Structure of mitochondria
- Dynamics of mitochondria
- Functions of mitochondria
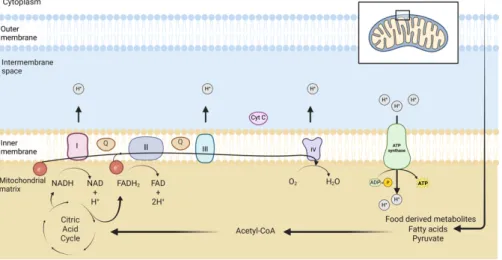
Interestingly, proteins involved in the replication, transcription and maintenance of the mitochondrial genome are nuclear encoded (Gustafsson et al., 2016). Oxygen molecules are the final electron acceptors in the respiratory chain (Alberts, 2008; Berg et al., 2012). The urea cycle is carried out in the cytosol and mitochondrial matrix (Berg et al., 2012).
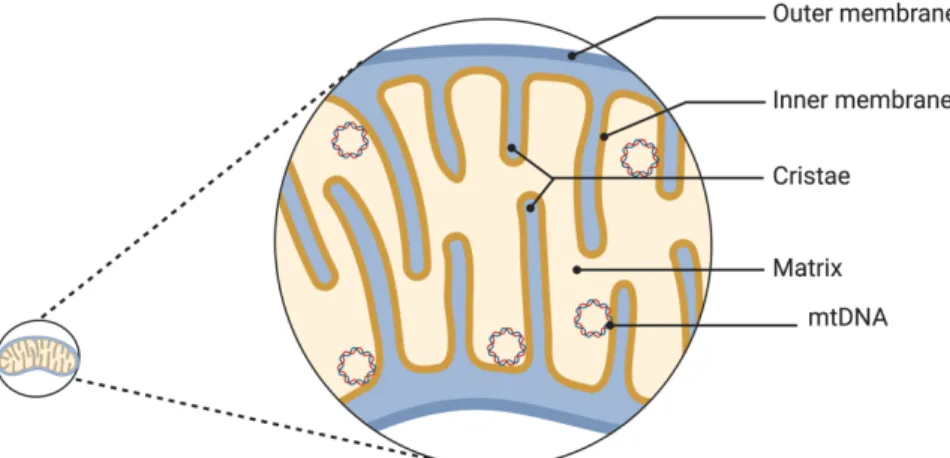
MITOCHONDRIAL GENOME
Human mitochondrial DNA
Iron in mitochondria is used for the synthesis of cofactors in iron-sulfur-containing proteins, heme group-containing proteins, and iron ion-containing proteins. These cofactors are essential for the proper functioning of proteins involved in oxidation-reduction reactions, DNA synthesis, repair, and other cellular processes (Paul et al., 2017). The mitochondrial genome encodes 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs), and 13 proteins that are essential components of OXPHOS complexes I, III, IV, and V ( Anderson et al., 1981 ; Gustafsson et al., 2016 ).
Genome organization and mitochondrial nucleoid
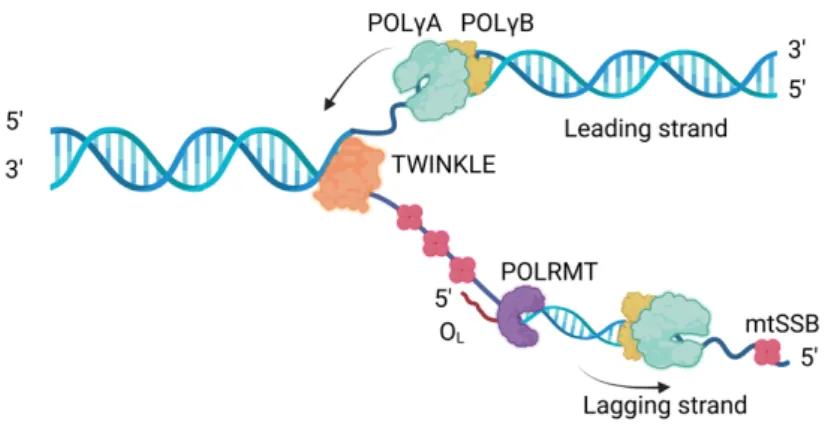
MITOCHONDRIAL DNA TRANSCRIPTION
Introduction
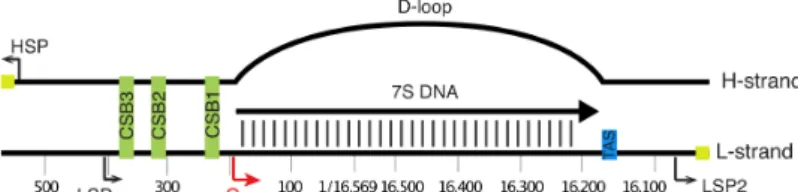
Recent studies using super-resolution microscopy have suggested that mammalian nucleoids contain approximately one mtDNA (Kukat et al., 2011). Nucleoids are associated with the IMM and they can also diffuse freely within the mitochondrial network (Brown et al., 2011). A second light strand promoter (LSP2) has recently been identified, but its physiological role remains to be investigated (Tan et al., 2022).
Mitochondrial transcription machinery
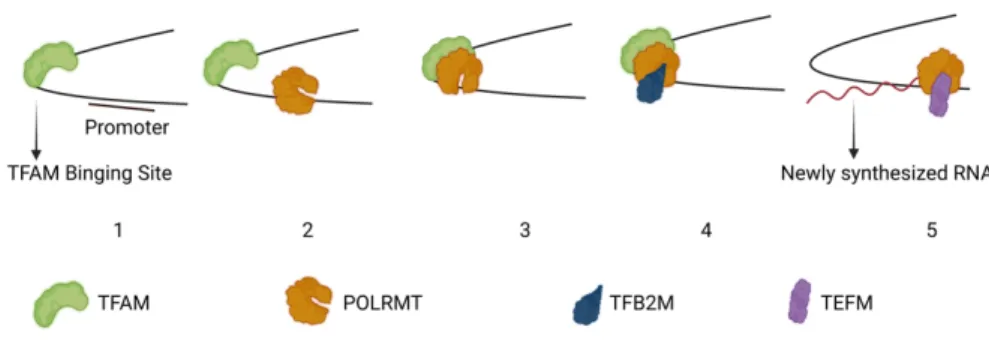
Transcription of mtDNA depends on nuclear-encoded transcription machinery that is completely different from the nuclear transcription machinery, but structurally similar to bacteriophage T7 (Tiranti et al., 1997; . Ringel et al., 2011). There are three essential factors required for complete mtDNA transcription: mitochondrial RNA polymerase (POLRMT), TFAM and mitochondrial transcription factor B2 (TFB2M) (Falkenberg et al., 2002; Posse & Gustafsson, 2017).
A model for mitochondrial DNA transcription
TFB2M was first discovered based on its sequence similarities to yeast mitochondrial transcription initiation factor (Mtf1) (Falkenberg et al., 2002; Sologub et al., 2009). Once it binds, TFAM induces a 180° turn in the DNA and recruits POLRMT to the promoter (Rubio-Cosials et al., 2011). The interaction between POLRMT and TFAM is mediated by the N-terminal extension (NTE) domain of POLRMT (Posse et al., 2014).
MITOCHONDRIAL DNA REPLICATION
- Introduction
- T7 DNA replication
- Mitochondrial replication machinery
- Mitochondrial nucleases
- The mode of mitochondrial DNA replication
- Mitochondrial diseases
It is a heterodimer composed of a catalytic subunit of POLgA and two additional POLgB subunits (Carrodeguas et al., 1999; Farge et al., 2007). Loss of exonuclease activity causes a significant increase in point mutations and premature aging phenotypes in mice (Trifunovic et al., 2004). MtSSB caps single-stranded DNA (ssDNA) during replication and prevents recombination of parental DNA strands and unwanted priming ( Miralles Fusté et al., 2014 ).
At its core, RNase H1 is involved in the maintenance of genome stability and telomere integrity, resolving R-loops (Arora et al., 2014; Parajuli et al., 2017). In mitochondria, RNase H1 plays a critical role in the maintenance of mtDNA in mice, as loss of RNase H1 causes embryonic lethality (Cerritelli et al., 2003). In addition, a study reported that RNase H1 is involved in the processing of mitochondrial ribosomal RNA precursors via interaction with mitochondrial protein P32 (Wu et al., 2013).
MGME1 is a mitochondrial exonuclease that shows preferential 3′-5′ activity on single-stranded DNA ( Kornblum et al., 2013 ; Szczesny et al., 2013 ). Furthermore, MGME1 was also shown to be implicated in the regulation of heavy strand replication and transcription termination (Matic et al., 2018). Depletion also causes single-strand breaks in mtDNA and ultimately induces apoptosis (Tann et al., 2011).
Mitochondrial dysfunction has also been associated with the biological aging process senescence (Trifunovic et al., 2004; Kauppila et al., 2017).
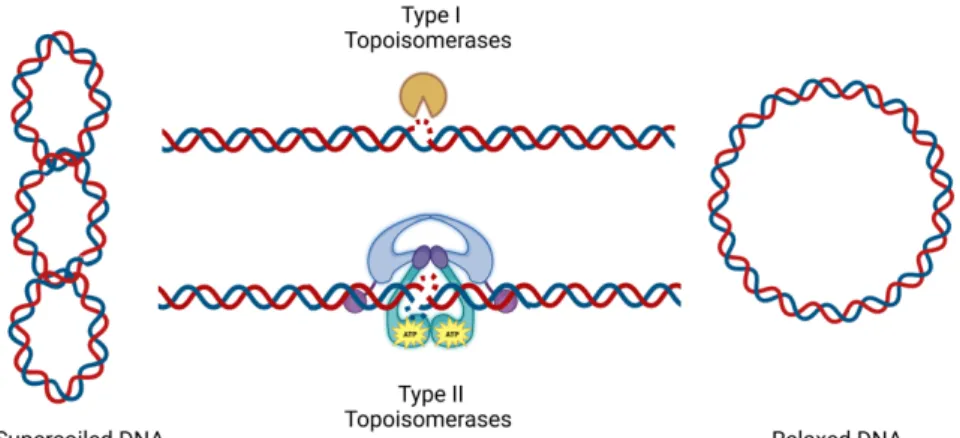
DNA TOPOLOGY
- Why do we need topoisomerases?
- Classification of topoisomerases
- Mitochondrial topoisomerases
- Topoisomerases and mitochondrial diseases
TOP2A is highly expressed in rapidly dividing cells such as pluripotent embryonic stem cells ( Thakurela et al., 2013 ). Furthermore, another study suggested that a truncated form of TOP2B was found in mitochondria (Low et al., 2003). However, more recent studies have failed to detect any of these type IIA topoisomerases in mitochondria (Nicholls et al., 2018; Menger et al., 2022).
However, more recent studies failed to detect any of these type IIA topoisomerases in mitochondria (Nicholls et al., 2018; Menger et al., 2022). Mitochondrial-specific TOP1MT has an N-terminal mitochondrial targeting sequence that facilitates its exclusive localization to mitochondria ( Dalla Rosa et al., 2009 ). TOP1MT directly interacts with POLRMT and transcriptionally active nucleoids containing POLRMT, removing transcription-associated supercoiling ( Sobek et al., 2013 ).
This suggests that TOP1MT has an important role in mtDNA hemostasis and hepatocyte proliferation (Khiati et al., 2015). Recently, the TOP3A protein was shown to be essential for mtDNA cleavage in mammalian cells (Nicholls et al., 2018). These termination structures are defined as hemicatenans, that is, double-stranded DNA (dsDNA) molecules linked through a single-stranded (ss) bond (Nicholls et al., 2018).
Mutations in the TOP3A gene cause mtDNA deletion, impaired mtDNA segregation, and ultimately mtDNA aggregation (Nicholls et al., 2018).
PAPER I
PAPER II
To gain a better understanding of the patient's phenotype, we performed not only structural modeling, but also a number of biochemical analyzes characterizing the activities of the mutant protein. We also analyzed the effects of the p.F907I mutation on the balance between proofreading and DNA synthesis activities, using a 3'-5' exonuclease and 5'-3' polymerase activity assay in the presence of increasing concentrations of dNTPs. To investigate the consequences of the p.F907I mutation in the context of the reconstituted replisome, we performed a rolling circle assay, which is a.
To overcome this disorder, we attempted to increase the concentrations of the mutant POLg, mtSSB or TWINKLE. To better understand the role of p.F907I, we constructed a replisome model using homology modeling based on the leading strand complex of the T7 bacteriophage replisome and POLg. However, crystallization experiments showed that the phenylalanine in T7 DNA polymerase gp5 corresponding to F907 is part of a surface-exposed hydrophobic pocket that tightly binds the C-terminal tail of the T7 gp4 helicase (Foster et al., 2019 ).
The majority of patients manifested with bilateral ptosis, ophthalmoplegia, myopathy and axonal sensory motor neuropathy, cerebellar ataxia, hearing loss and cardiac abnormalities. We first performed a modeling of mutations in the TOP3A structure, which revealed that most of the mutations cluster in the Toprim domain (topoisomerase primase), with few exceptions. In the ssDNA binding assay, we found that except for two variants, all other patient variants showed binding activity similar to that of the WT protein.
Based on our results, we propose that the level of catalytic impairment of the enzyme determines whether TOP3A mutations manifest as mitochondrial disease or Bloom syndrome.
PAPER III
To investigate the effects of TOP3A mutations associated with impaired mitochondrial genome stability, we used a number of biochemical assays and monitored several aspects of protein function, such as ssDNA binding, unwinding of supercoiled DNA molecules, and decatenation of ssDNA substrates. As mitochondrial TOP3A is involved in the separation of the products of hemicatenated mtDNA replication (Nicholls et al., 2018), we attempted to mimic the in vivo activity of TOP3A using ssDNA catenase substrates with a bond number of 1, implying that a single catalytic cycle of TOP3A can resolve this. Based on our findings, we propose that the degree of catalytic defect of the enzyme determines whether TOP3A mutations manifest as mitochondrial disease or Bloom syndrome. primates), with some exceptions.
Since mitochondrial TOP3A is involved in the disassembly of hemicatenate mtDNA replication products (Nicholls et al., 2018), we attempted to mimic the in vivo activity of TOP3A using ssDNA catenane substrates with a coupling number of 1, meaning, that a single catalytic cycle of TOP3A can resolve it. Mitochondrial localization of TOP3A and TOP1MT is supported by bioinformatic methods such as MitoCarta 2.0 and MitoMiner. To further investigate the subcellular localization of the six human topoisomerases, we used super-resolution Airyscan confocal microscopy.
We found that full-length TOP3A showed exclusively mitochondrial localization, whereas the shorter TOP3A isoform lacking the first 25 amino acids (representing the mitochondrial targeting sequence) localized exclusively to the nucleus. We monitored the pattern of mtDNA replication in TOP3A- and TOP1MT-depleted cells using 2DNAGE. To further analyze the contribution of TOP3A and TOP1MT to mtDNA replication, whole genome sequencing and Southern blot.
To confirm the contribution of both topoisomerases to mtDNA transcription, we performed an in vitro transcription reaction using supercoiled template and the transcription machinery proteins POLRMT, TFAM, TFB2M and TEFM in the absence or presence of TOP3A and TOP1MT.
PAPER IV
EXOG was highly active on RNA, even in the presence of the LRPPRC/SLIRP complex. The importance of the mitochondrial genome for human biology has been increasingly recognized in recent decades, especially after the discovery of an increasing number of human diseases associated with mutations in the mtDNA itself or protein components that help maintain the mitochondrial genome. For example, our studies of the underlying pathogenic mechanisms associated with the phenylalanine to isoleucine mutation at position 907 of human POLγA have shown that POLg can form transient interactions with TWINKLE at mitochondrial replication forks.
The p.F907I mutation did not affect basic activities of POLg, including 3¢-5¢ exonuclease activity or DNA polymerization, on an ssDNA template, but failed to operate on dsDNA in the context of the mitochondrial replisome . Our studies suggest that the overall severity of the TOP3A mutations determines the clinical presentations. The accessory subunit of Xenopus laevis Mitochondrial DNA polymerase γ Increases the processivity of the catalytic subunit of human DNA polymerase γ and is related to Class II Aminoacyl-tRNA synthetases.
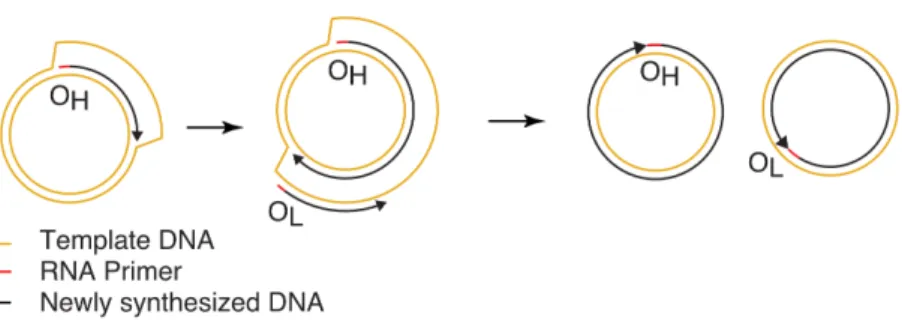
Mitochondrial RNA polymerase is required for activation of the light-strand DNA replication origin. Two-dimensional DNA agarose electrophoresis of intact mitochondrial DNA reveals the structural complexity of the mammalian mitochondrial genome. Human mitochondrial transcription revisited: Only TFAM and TFB2M are required for mitochondrial gene transcription in vitro.
Homologues of mitochondrial transcription factor B, sparsely distributed within the eukaryotic radiation, are likely derived from the dimethyladenosine methyltransferase of the mitochondrial endosymbiont. BLAP18/RMI2, a novel OB fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Identification of the gene encoding the human mitochondrial RNA polymerase (h-mtRPOL) by cyberscreening the Expressed Sequence Tags database.