Iranian Journal of Basic Medical Sciences
ijbms.mums.ac.ir
Association of ADAM33 gene polymorphisms with allergic
asthma
Mohammad Reza Zand Karimi
1#, Reza Faridhosseini
1#, Mohammad Reza Abbaszadegan
2,
Farrahzad Jabbari azad
1, Afshin Shirkani
4, Anali Riyahi
2,
Mehdi Montazar
3,
Mehran Gholamin
2*
1 Allergy Research Center, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran2 Division of Human Genetics, Immunology Research Center, Avicenna Research Institute, Mashhad University of Medical Sciences, Mashhad, Iran
3 Department of Pathology, Omid Hospital, Mashhad University of Medical Sciences, Mashhad, Iran
4 Bushehr University of Medical Science, School of Medicine. Allergy and clinical Immunology. Department, Bushehr, Iran # Contributed equally to this work
A R T I C L E I N F O A B S T R A C T
Article type:
Original article Objective(s): Asthma results from the interaction between genetic and environmental factors. ADAM33 gene on chromosome 20p13 is associated with asthma and airway hyperresponsiveness. Materials and Methods: This is a case-control study, where four SNPs S1 (rs3918396), T1 (rs2280091), T2 (rs2280090), V4 (rs2787094) of ADAM33 gene have been assessed in patients with allergic asthma and normal controls (95 patients and 86 normal). Blood samples of these participants have been genotyped by PCR and the RFLP method.
Results: There was no association between asthmatic patients and polymorphisms of alleles, genotypes and haplotypes of the ADAM33 gene. When categorizing the asthmatic patients in severe, moderate and mild groups, associations in the subcategories of asthmatic patients were found. There were associations between polymorphisms of C allele of T1 SNP with severe asthmatic patients and G allele of V4 SNP with moderate asthmatics respectively (P=0.006,
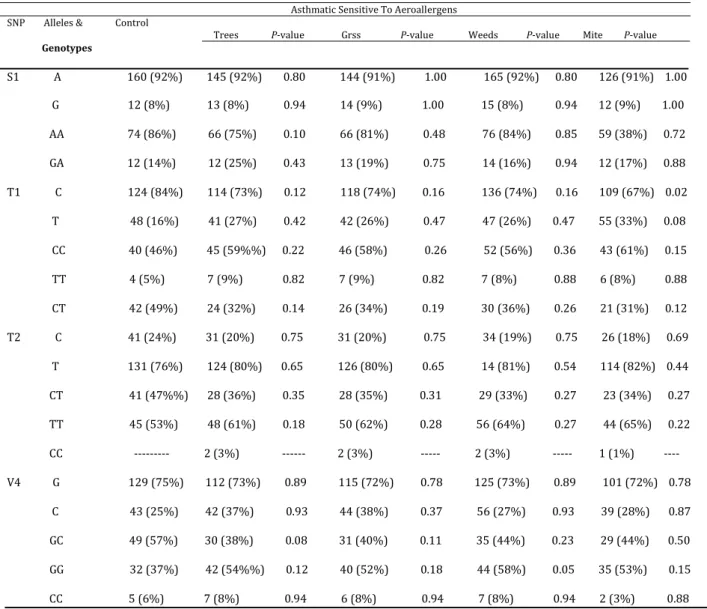
P=0.01). There was a significant association between sensitivity to mite and polymorphism of C allele of T1 SNP (P=0.02). Besides, there was a significant association between sensitivity to weeds and genotype GG of V4 SNP (P=0.05).
Conclusion: Polymorphisms of ADAM33 gene might be associated with severe asthma and sensitivity to aeroallergens in northeast of Iran, but further studies are needed to determine the polymorphisms in this area and other regions of our country.
Article history: Received: Nov 12, 2013 Accepted: Jan 28, 2014
Keywords: ADAM33 Allergic asthma Genetic SNP
►
Please cite this paper as:Zand Karimi MR, Faridhosseini R, Abbaszadegan MR, Jabbari azad F, Shirkani A, Riyahi A, Montazar M, GholaminM. Association of ADAM33 gene single nucleotide polymorphisms with allergic asthma in northeastern Iranian population. Iran J Basic Med Sci 2014; 17:716-721.
Introduction
Asthma is a worldwide problem that is more common in developed countries. An estimated 300 million people worldwide are suffering from asthma. The prevalence of asthma in children and adults is 3 to 38 and 2 to 12 percent, respectively. Asthma affects 10% of people in Iran. In recent decades, the prevalence of asthma has increased especially in developed countries
(1–3). Asthma is a chronic lung diseasecharacterizedby
increased bronchial responsiveness, obstructionofthe
airways and pulmonary symptoms. In this disease, acute and chronic inflammation and tissue changes include a thickened airway, sub-epithelial fibrosis and increased smooth muscle mass. Although the majority of people who are suffering from asthma have normal lung function, accelerated loss of lung function can
occur in somepeople(4–6). Asthma results from the
interaction of genetic and environmental factors. 20p13 chromosome is associated with asthma and increased airway responsiveness. A disintegrin and metallopro-tease domain 33 (ADAM33) gene in this chromosomal region is a candidate gene for asthma and increased bronchial responsiveness. This gene is one of the members of the metalloproteinase and disintegrin protein family that is classified as disintegrin and metalloproteina- ses (7). ADAM family includes three members with very complex structures. ADAM proteins are transmembrane proteins with multiple domains, which are involved in many cellular activities. ADAM family plays an important role in cell adhesion,
proliferation, differentiation, signal transduction,
apoptosis and inflammatory responses. This gene is
expressed in lung fibroblasts and smooth muscle
and it has been associated with increased bronchial
Table 1. Features of SNP, primer sequence, length of PCR product and restriction enzyme
Name of SNP Primer sequence PCR product (b.p.) Restriction enzyme
S1 (rs3918396) Forward: 5′-TGTGCAGGCTGAAAGTATGC-3'
Reverse: 5′-AGAGCTCTGAGGAGGGGAAC-3'
304 BtsCI
T1 (rs2280091) Forward: 5'-ACTCAAGGTGACTGGGTGCT-3' Reverse: 5'-GAGGGCATGAGGCTCACTTG-3
400 NcoI
(Bsp19I)
T2 (rs2280090) Forward: 5'-TTCTCAGGGTCTGGGAGAAA-3'
Reverse: 5'-GCCAACCTCCTGGACTCTTA-3'
310 HpyCH4III
(TaaI) V4 (rs2787094) Forward: 5'-ACACACAGAATGGGGGAGAG-3'
Reverse: 5'-CCAGAAGCAAAGGTCACACA-3'
374 PstI
(BstMAI)
responsiveness (8–11).The present study was designed
to investigate the associationbetweenfour ADAM33
gene polymorphisms and allergic asthma by PCR-RLFP method.
Materials and Methods
SubjectsThe participants, who were referred to Ghaem Hospital, filled out a questionnaire for identifying their signs and symptoms according to Global Initiative for Asthma (GINA) guidelines. After obtaining informed consent from the patients, spirometry was performed for each patient based on forced expiratory volume in 1 sec (FEV1), forced vital capacity (FVC) and the ratio FEV1/FVC in either normal situation or 10-15 min after exercise and/or bronchodilator administration. Skin allergy prick test was done for diagnosis of allergy in all participants. Allergen extracts used in the skin prick test included extracts of common allergens consisting of aeroallergens (pollens of trees, grasses and weeds) and dust mites. After providing an informed consent, the control group filled out a questionnaire. Then, clinical examination and the prick skin test were performed for allergic evaluations.
Genotyping
Five milliliters of venous blood obtained from each participant was collected in falcon containing EDTA. Genomic DNA was extracted using DNA extraction kit (Pars Toos,Iran). Using specific primers for the four SNP types (T1, T2, S1, and V4), PCR was performed, then, SNP types in amplified fragment were evaluated using restriction enzymes. Features of the SNPs, primers sequence, PCR product length and restriction enzymes are shown in Table 2.
Statistics
Frequency of allele and genotypes between case and control groups and individual subgroups were analyzed by X2 test. Statistical analysis was performed using the SPSS software (Version 16.0).
The P-values equal to or less than 0.05 are
considered as significant. Statistical analysis of haplotype was performed by Epi. Info software. The distribution of allele frequencies was tested for
the χ test. The degree of linkage disequilibrium
between loci was measured using Lewontin’s Dm.
Results
The mean ages of the asthma patients and the control group were 34 years (range, 7 to 64 years), and
29 years (range, 2 to 75 years), respectively. 66% of
patients and 37% of the control subjects were female and 29 patients (30%) had a family history of asthma. In the case group, 75 (79%), 75(79%), 84 (88%), and 70 (74%) of patients were sensitive to the tree pollen, grass, weeds, and mite, respectively. The demographics of patients and genotype frequencies in the asthma and control groups are listed in Tables 2 and 3, respectively.
Characterization of alleles and genotypes of different types of phenotypes in patients with asthma is presented in Table 6. There was an association between polymorphism of C allele of T1 SNP with severe asthma
(P=0.006). Also, a significant difference was found
between G allele of V4 SNP with moderate asthmatics
(P=0.01). There was no association between asthmatic
patients and polymorphism of ADAM33 gene and other phenotypes.
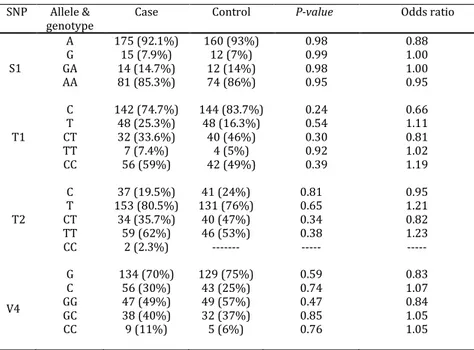
Frequency of alleles and genotypes of four SNPs (S1, T1, T2, and V4) are presented in Table 4. There were no associations between alleles and genotypes of the four SNPs in the patient group when compared to the control group.
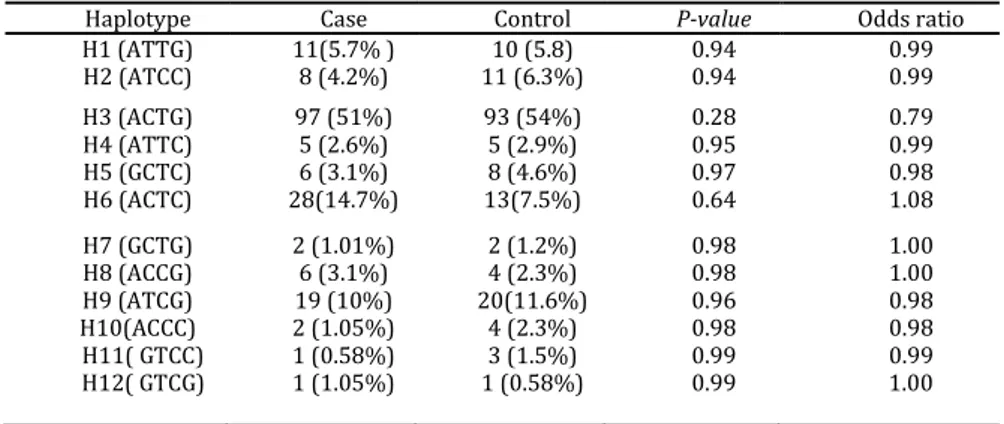
Different haplotypes of ADAM33 gene are presented in Table 5. There were no significant differences between 12 types of haplotypes in the two groups. There was a significant association between sensitivity to mite and polymorphism of C allele of
T1 SNP (P=0.02). Besides, there was a significant
association between sensitivity to weeds and
genotype GG of V4 SNP (P=0.05). There was no
significant association among other alleles and genotypes as shown in Table 7.
Table 2. The Hardy–Weinberg equilibrium analysis for case and control
SNP genotypes types
HWE Con (P-value)
HWE Case (P-value)
HWE Case/Con (P-value)
S1 0.00 0.74 0.00
T1 0.35 0.72 0.89
Table 3. Demographic characterization of patients and control group
Variable Control group (n=86) Patients (n=95)
Sex: Male Female
49(57%) 37 (43%)
42 (44%) 53 (56%) Age (year):
Minimum Maximum Intermediate
2 75 39
7 64 34 Maximum capacity of expiratory:
Minimum Maximum Intermediate
─ ─ ─
34% 128%
81%
Maximum expiratory volume in 1st sec: Minimum
Maximum
Intermediate ──
─
30% 124%
78% Severity of asthma:
mild moderate severe
─ ─ ─
52 (54%) 28 (30%) 15 (15%)
Family history of asthma ─ 29 (30%)
Prick skin test Tree pollen grass weeds mite
─ ─ ─ ─
75 (79%) 75 (79%) 84 (88%) 70 (74%)
Table 4. Frequency of alleles and genotypes of ADAM33 in patients with asthma and the control group
SNP Allele & genotype
Case Control P-value Odds ratio
S1
A G GA AA
175 (92.1%) 15 (7.9%) 14 (14.7%) 81 (85.3%)
160 (93%) 12 (7%) 12 (14%) 74 (86%)
0.98 0.99 0.98 0.95
0.88 1.00 1.00 0.95
T1 C T CT TT CC
142 (74.7%) 48 (25.3%) 32 (33.6%) 7 (7.4%) 56 (59%)
144 (83.7%) 48 (16.3%) 40 (46%)
4 (5%) 42 (49%)
0.24 0.54 0.30 0.92 0.39
0.66 1.11 0.81 1.02 1.19
T2 C T CT TT CC
37 (19.5%) 153 (80.5%) 34 (35.7%) 59 (62%)
2 (2.3%)
41 (24%) 131 (76%) 40 (47%) 46 (53%) ---
0.81 0.65 0.34 0.38
---0.95 1.21 0.82 1.23 ---
V4
G C GG GC CC
134 (70%) 56 (30%) 47 (49%) 38 (40%) 9 (11%)
129 (75%) 43 (25%) 49 (57%) 32 (37%) 5 (6%)
0.59 0.74 0.47 0.85 0.76
0.83 1.07 0.84 1.05 1.05
Table 5. Frequency of haplotypes of ADAM33 in patients with asthma and the control group
Haplotype Case Control P-value Odds ratio
H1 (ATTG) 11(5.7% ) 10 (5.8) 0.94 0.99
H2 (ATCC) 8 (4.2%) 11 (6.3%) 0.94 0.99
H3 (ACTG) 97 (51%) 93 (54%) 0.28 0.79
H4 (ATTC) 5 (2.6%) 5 (2.9%) 0.95 0.99
H5 (GCTC) 6 (3.1%) 8 (4.6%) 0.97 0.98
H6 (ACTC) 28(14.7%) 13(7.5%) 0.64 1.08
H7 (GCTG) 2 (1.01%) 2 (1.2%) 0.98 1.00
H8 (ACCG) 6 (3.1%) 4 (2.3%) 0.98 1.00
H9 (ATCG) 19 (10%) 20(11.6%) 0.96 0.98
H10(ACCC) 2 (1.05%) 4 (2.3%) 0.98 0.98
H11( GTCC) 1 (0.58%) 3 (1.5%) 0.99 0.99
H12( GTCG) 1 (1.05%) 1 (0.58%) 0.99 1.00
Table 6. Frequency of alleles and genotypes of ADAM33 in patients with asthma and the control group
Alele & genotype Control Asthmatic patients
S1 A
G AA GA 160 (93%) 12 (7%) 74 (86%) 12 (14%)
Mild P-value Moderate P-value Severe P-value
95 (91%) 9 (9%) 43 (83%) 9 (17%) 0.86 0.94 0.72 0.88 52 (93%) 4 (7%) 24 (86%) 4 (14%) 0.99 0.94 0.85 0.94 28 (93%) 2 (7%) 13 (87%) 2 (13%) 0.79 0.94 1.00 1.00
T1 C
T CC TT CT 144 (84%) 48 (16%) 40 (46%) 4 (5%) 42 (49%) 80 (77%) 24 (23%) 30 (59%) 3 (6%) 18 (35%) 0.33 0.63 0.22 1.00 0.22 44 (76%) 14 (24%) 18 (64%) 2 (8%) 8 (28%) 0.27 0.58 0.07 0.88 0.07 19 (63%) 11 (27%) 5 (33%) 1 (7%) 9 (60%) 0.006 0.09 0.27 0.94 0.77
T2 C
T CT TT CC 41 (24%) 131 (76%) 41 (47%) 45 (53%) ---20 (19%) 84 (81%) 18 (35%) 33 (63%) 1 (2%) 0.75 0.54 0.31 0.22 9 (16%) 47 (84% 9 (34%) 19 (66%) --- 0.58 0.21 0.23 0.18 --- 9 (30%) 21 (70%) 9 (38%) 6 (24%) --- 0.68 0.49 0.45 0.38 ---
V4 G
C GG GC CC 129 (75%) 43 (25%) 49 (57%) 32 (37%) 5 (6%) 76 (73%) 28 (27%) 28 (54%) 20 (38%) 4 (8%) 0.89 0.93 0.83 1.00 0.94 37 (54%) 32 (46%) 11 (40%) 15 (55%) 2 (5%) 0.01 0.08 0.11 0.10 1.00 21 (70%) 9 (30%) 9 (60%) 3 (20%) 3 (20%) 0.59 0.74 0.74 0.18 0.32
Discussion
Van Eerdewegh reported that the ADAM33 gene is expressed restrictively in human fibroblasts and smooth muscles. ADAM33 polymorphism can accelerate proliferation in the smooth muscles and fibroblasts, which causes sub-epithelial fibrosis and increased bronchial responsiveness. ADAM33 gene, with specific polymorphisms increases shift toward inflammation or immune responses mediated by the
T-helper 2 cell (11–13).
Several studies have found a significant difference in ADAM33 gene expression between the healthy and asthmatic subjects. For example, Van Eerdewegh observed a significant correlation between seven SNP alleles (F+1, Q-1, S1, S2, ST+4, V-1, and V4) in 421 families (6). Moreover, similar results were reported by Robby Benjamin in America, and by Jay H Lee and LG
Bloki in Korea (7–9). In the present study the
relationship among ADAM33 gene polymorphisms, the severity of asthma and allergic reactions was investigated. This case-control study was performed on
northeast of Iran, Mashhad. We evaluated the four SNPs that more studies have focused on. This investigation was done for the first time in Iran. Our results showed that there was no significant association between the asthma patients and the control group with regard to ADAM33 gene polymorphisms and haplotypes. In line with Elçin Bora in Turkey, Denise L. Lind in Puerto Rico, and Wang, P. Liu in China, we found no relationship between ADAM33 gene polymorphisms and asthma
(14–16).
However, several studies have reported a signify-cant relationship between ADAM33 gene polymer-phisms, asthma, increased bronchial responsiveness to methacholine, restrictive abnormal airway and total serum IgE levels in America, England, Germany, China, Japan, South Korea, Latin America, India,
Australia, Turkey and Saudi Arabia (17–30). In the
Table 7. Investigation of gene polymorphism in patients with allergic asthma and the control group
Asthmatic Sensitive To Aeroallergens SNP Alleles & Control
Trees P-value Grss P-value Weeds P-value Mite P-value
Genotypes
S1 A 160 (92%) 145 (92%) 0.80 144 (91%) 1.00 165 (92%) 0.80 126 (91%) 1.00
G 12 (8%) 13 (8%) 0.94 14 (9%) 1.00 15 (8%) 0.94 12 (9%) 1.00
AA 74 (86%) 66 (75%) 0.10 66 (81%) 0.48 76 (84%) 0.85 59 (38%) 0.72
GA 12 (14%) 12 (25%) 0.43 13 (19%) 0.75 14 (16%) 0.94 12 (17%) 0.88
T1 C 124 (84%) 114 (73%) 0.12 118 (74%) 0.16 136 (74%) 0.16 109 (67%) 0.02
T 48 (16%) 41 (27%) 0.42 42 (26%) 0.47 47 (26%) 0.47 55 (33%) 0.08
CC 40 (46%) 45 (59%%) 0.22 46 (58%) 0.26 52 (56%) 0.36 43 (61%) 0.15
TT 4 (5%) 7 (9%) 0.82 7 (9%) 0.82 7 (8%) 0.88 6 (8%) 0.88
CT 42 (49%) 24 (32%) 0.14 26 (34%) 0.19 30 (36%) 0.26 21 (31%) 0.12
T2 C 41 (24%) 31 (20%) 0.75 31 (20%) 0.75 34 (19%) 0.75 26 (18%) 0.69
T 131 (76%) 124 (80%) 0.65 126 (80%) 0.65 14 (81%) 0.54 114 (82%) 0.44
CT 41 (47%%) 28 (36%) 0.35 28 (35%) 0.31 29 (33%) 0.27 23 (34%) 0.27
TT 45 (53%) 48 (61%) 0.18 50 (62%) 0.28 56 (64%) 0.27 44 (65%) 0.22
CC --- 2 (3%) --- 2 (3%) --- 2 (3%) --- 1 (1%) ----
V4 G 129 (75%) 112 (73%) 0.89 115 (72%) 0.78 125 (73%) 0.89 101 (72%) 0.78
C 43 (25%) 42 (37%) 0.93 44 (38%) 0.37 56 (27%) 0.93 39 (28%) 0.87
GC 49 (57%) 30 (38%) 0.08 31 (40%) 0.11 35 (44%) 0.23 29 (44%) 0.50
GG 32 (37%) 42 (54%%) 0.12 40 (52%) 0.18 44 (58%) 0.05 35 (53%) 0.15
CC 5 (6%) 7 (8%) 0.94 6 (8%) 0.94 7 (8%) 0.94 2 (3%) 0.88
There was no genetic correlation in the remaining alleles, and no significant differences between values of alleles and genotypes in patients with asthma. Besides, there was a significant difference between values of the SNP T1 allele in patients with severe asthma compared to the control ones. It is highly probable that this association is more significant if a larger population is studied. Furthermore, it has been shown that there is an association between moderate asthma and the V4 polymorphism. On the
other hand the Hardy–Weinberg equilibrium is
present in this group. There was a relationship between allergic sensitivity to aeroallergens and ADAM33 gene polymorphism in patients suffering from asthma. Similar studies have investigated the association between gene polymorphism and total IgE levels, but a study using prick skin test is unprecedented in the literature. Hence, our study is the first to investigate the relationship between ADAM33 gene polymorphism and allergic sensitivity
northeast of Iran; since Iran has diverse ethnic groups, it is suggested to perform similar studies on other populations. Polymorphisms of ADAM33 gene might be associated with severe asthma and sensitivity to aeroallergens.
Conclusion
Polymorphisms of ADAM33 gene might be associated with severe asthma and sensitivity to aeroallergens in the northeastern population of Iran. Further studies are needed in other districts of Khorasan and Iran.
Acknowledgment
References
1. Sharma N, Tripathi P, Awasthi S. Role of ADAM33 gene and associated single nucleotide polymorphisms in asthma. Allergy Rhinol ;2011;2:63-70.
2.Entezari A, Mehrabi Y, Varesvazirian M, Pourpak Z, Moin M. A systematic review of recent asthma symptom surveys in Iranian children. Chron Respir Dis 2009;6:109- 14.
3.Al-Khayyat AI, Al-Anazi M, Warsy A, Vazquez-Tello A, Alamri AM, Halwani R, et al. T1 and T2 ADAM33 single nucleotide polymorphisms and the risk of childhood asthma in a Saudi Arabian population: a pilot study. Ann Saudi Med;2012;32:479-486. 4. Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, et al. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy 2004; 34:757-760.
5. Vercelli D. Discovering susceptibility genes for asthma and allergy. Nat Rev Immunol 2008; 8:169-182. 6. Haitchi HM, Powell RM, Shaw TJ, Howarth PH, Wilson SJ, Wilson DI, et al. ADAM33 expression in asthmatic airways and human embryonic lungs. Am J Respir Crit Care Med 2005 ; 171:958-965.
7. Kedda MA, Duffy DL, Bradley B, O'Hehir RE, Thompson PJ. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet 2006; 14:1027-1036.
8. Schedel M, Depner M, Schoen C, Weiland SK, Vogelberg C, Niggemann B, et al. The role of polymorphisms in ADAM33, a disintegrin and metalloprotease 33, in childhood asthma and lung function in two German populations. Respir Res 2006; 7:91.
9. Tripathi P, Awasthi S, Prasad R, Husain N, Ganesh S. Association of ADAM33 gene polymorphisms with adult-onset asthma and its severity in an Indian adult population. J Genet; 2011; 90:265-273.
10. Awasthi S, Tripathi P, Ganesh S, Husain N. Association of ADAM33 gene polymorphisms with asthma in Indian children. J Hum Genet; 2011; 56:188-195.
11. Holgate ST, Yang Y, Haitchi HM, Powell RM, Holloway JW, Yoshisue H, et al. The genetics of asthma: ADAM33 as an example of a susceptibility gene. Proc Am Thorac Soc 2006 ; 3:440-443.
12. Van Eerdewegh P, Little RD, Dupuis J, Del Mastro RG, Falls K, Simon J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature 2002 ; 418:426-430.
13. Howard TD, Postma DS, Jongepier H, Moore WC, Koppelman GH, Zheng SL, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol 2003; 112:717-722.
14. Elçin Bora ZA-A, Fatih Firinci, Tufan Çankaya, Özlem Giray-Bozkaya, Nevin Uzuner, Ülgenalp A. ADAM33 Gene Polymorphisms Are Not Associated with Asthma in Turkish Children Pediatric Allergy, Immunology, and Pulmonology. 2012; 25(2): 97-100. 15. Lind DL, Choudhry S, Ung N, Ziv E, Avila PC, Salari K, et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir
16. Wang P, Liu QJ, Li JS, Li HC, Wei CH, Guo CH, et al. Lack of association between ADAM33 gene and asthma in a Chinese population. Int J Immunogenet 2006; 33:303-306.
17. Sadeghnejad A, Ohar JA, Zheng SL, Sterling DA, Hawkins GA, Meyers DA, et al. Adam33 polymorphisms are associated with COPD and lung function in long-term tobacco smokers. Respir Res 2009; 10:21.
18. Raby BA, Silverman EK, Kwiatkowski DJ, Lange C, Lazarus R, Weiss ST. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol 2004 ; 113:1071-1078.
19. Lee JH, Park HS, Park SW, Jang AS, Uh ST, Rhim T, et al. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy 2004 ; 34:860-865.
20. Blakey J, Halapi E, Bjornsdottir US, Wheatley A, Kristinsson S, Upmanyu R, et al. Contribution of ADAM33 polymorphisms to the population risk of asthma. Thorax 2005; 60:274-276.
21. Li X, Zhang Y, Zhang J, Xiao Y, Huang J, Tian C, et al. Asthma susceptible genes in Chinese population: a meta-analysis. Respir Res; 2010;11:129.
22. Werner M, Herbon N, Gohlke H, Altmuller J, Knapp M, Heinrich J, et al. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy 2004; 34:26-31.
23. Simpson A, Maniatis N, Jury F, Cakebread JA, Lowe LA, Holgate ST, et al. Polymorphisms in a disintegrin and metalloprotease 33 (ADAM33) predict impaired early-life lung function. Am J Respir Crit Care Med 2005; 172:55-60.
24. van Diemen CC, Postma DS, Vonk JM, Bruinenberg M, Schouten JP, Boezen HM. A disintegrin and metalloprotease 33 polymorphisms and lung function decline in the general population. Am J Respir Crit Care Med 2005; 172:329-333.
25. Foley SC, Mogas AK, Olivenstein R, Fiset PO, Chakir J, Bourbeau J, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol 2007; 119:863-871. 26. Hirota T, Hasegawa K, Obara K, Matsuda A, Akahoshi M, Nakashima K, et al. Association between ADAM33 polymorphisms and adult asthma in the Japanese population. Clin Exp Allergy 2006; 36:884-891.
27. Thongngarm T, Jameekornrak A, Limwongse C, Sangasapaviliya A, Jirapongsananuruk O, Assawamakin A, et al. Association between ADAM33 polymorphisms and asthma in a Thai population. Asian Pac J Allergy Immunol 2008; 26:205-211.
28. Bijanzadeh M, Ramachandra NB, Mahesh PA, Mysore RS, Kumar P, Manjunath BS, et al. Association of IL-4 and ADAM33 gene polymorphisms with asthma in an Indian population. Lung; 2010; 188:415-422.
29. El-Falaki M WM, Ezzat G, Mokhtar D, El-Baz M, Hamed D. A disintegrin and metalloproteinase 33 (ADAM33) gene polymorphism association with asthma in Egyptian children. Egyp J Med Hum Genet 2013; 14:55-62.