The long non-coding RNA ADAMTS9-AS2 was identified as a potential
prognostic biomarker in triple-negative breast cancer
O RNA longo não codificador ADAMTS9-AS2 foi identificado como um
potencial biomarcador prognóstico no câncer de mama triplo negativo
DOI:10.34117/bjdv6n3-479
Recebimento dos originais: 10/02/2020 Aceitação para publicação: 30/03/2020
Daniel Rodrigues de Bastos
Discipline of Oncology, Department of Radiology and Oncology, Faculty of Medicine, University of São Paulo, 01246-903, São Paulo, Brazil;
Laboratory of Molecular Genetics, Center for Translational Research in Oncology, Cancer Institute of São Paulo, 01246-000, São Paulo, Brazil.
danielbastos.adm@gmail.com
Jean Michel Rocha Sampaio Leite
Faculty of Public Health – FSP, University of São Paulo (USP)
Mércia Patrícia Ferreira Conceição
Discipline of Oncology, Department of Radiology and Oncology, Faculty of Medicine, University of São Paulo, 01246-903, São Paulo, Brazil;
Laboratory of Molecular Genetics, Center for Translational Research in Oncology, Cancer Institute of São Paulo, 01246-000, São Paulo, Brazil.
Cesar Augusto Sam Tiago Vilanova-Costa
Laboratory of Tumoral Biology and Oncogenetics, Araujo Jorge Hospital, Association to Combat Cancer of Goias, Goiania, GO, Brazil
Antonio Márcio Teodoro Cordeiro Silva
School of Medical Sciences, Biomedics and Pharmaceuticals, Departament of Medicine, Pontifical Catholic University of Goiás, Goiania, GO, Brazil
Maria Aparecida Nagai
Discipline of Oncology, Department of Radiology and Oncology, Faculty of Medicine, University of São Paulo, 01246-903, São Paulo, Brazil;
Laboratory of Molecular Genetics, Center for Translational Research in Oncology, Cancer Institute of São Paulo, 01246-000, São Paulo, Brazil.
ABSTRACT
Introduction: Breast cancer is the main cause of cancer-related deaths in women in Brazil and around the world. Triple-negative breast cancer (TNBC) subtype is a complex disease that has a poor prognosis and limited therapeutic options, being chemotherapy the mainstay of treatment for the patients. A variety of long non-coding RNA was associated with tumoral progression and chemoresistance in breast cancer. Objective: The aim of this study was to identify a potential prognostic biomarker in TNBC patients. Methods: A microarray dataset containing expression signatures of 165 TNBC (GSE76250) was evaluated, and an in silico analysis was conducted to identify new possible biomarkers in TNBC. Results: ADAMTS9-S2 was the most significant
transcript identified by GEO2R tools. Low levels of ADAMTS9-S2 were observed in hormone receptors negative (p<0.0001) and high expression in HER2-negative patients (p<0.0001). In our study, low expression of ADAMTS9-S2 was associated poor relapse-free survival in TNBC patients. Conclusion: Our study provides evidence that ADAMTS9-S2 may be a potential prognostic biomarker in TNBC patients. However, further functional studies are necessary to understand, characterize, and confirm the ADAMTS9-AS2 expression patterns and relation with TNBC progression.
Keywords:lncRNA, breast cancer, biomarker, TNBC. RESUMO
Introdução: O câncer de mama é a principal causa de morte relacionada ao câncer em mulheres no Brasil e no mundo. O subtipo de câncer de mama triplo negativo (TNBC) é uma doença complexa, com prognóstico ruim e opções terapêuticas limitadas, sendo a quimioterapia a base do tratamento para as pacientes. Uma variedade de RNA longos não codificantes foram associados à progressão tumoral e à quimiorresistência no câncer de mama. Objetivo: O objetivo deste estudo foi identificar um potencial biomarcador prognóstico em pacientes com TNBC. Métodos: Um conjunto de dados de microarranjos contendo assinaturas de expressão de 165 TNBC (GSE76250) foi avaliado e uma análise in silico foi realizada para identificar novos possíveis biomarcadores. Resultados: ADAMTS9-S2 foi o transcrito mais significativo identificado pela ferramenta GEO2R. Baixos níveis de ADAMTS9-S2 foram observados em receptores hormonais negativos (p <0,0001) e alta expressão em pacientes com HER2 negativo (p <0,0001). Em nosso estudo, a baixa expressão de ADAMTS9-S2 foi associada à pior sobrevida livre de recidiva em pacientes com TNBC. Conclusão: Nosso estudo fornece evidências de que o ADAMTS9-S2 pode ser um potencial biomarcador prognóstico em pacientes com TNBC. No entanto, estudos funcionais são necessários para entender, caracterizar e confirmar os padrões de expressão do ADAMTS9-AS2 e a relação com a progressão do TNBC.
Palavras-chave: lncRNA, câncer de mama, biomarcador, TNBC. 1 INTRODUCTION
Breast cancer is the leading cause of cancer-related deaths in women in Brazil and around the world (1). In spite of the considerable advances made, mortality figures are still alarming. Among the factors significantly contributing to these numbers, tumor heterogeneity that correlates to immune response, tumor progression, and therapeutic resistance against current treatments are highly relevant (2).
Triple-negative Breast Cancer (TNBC), based on immunohistochemistry (IHC), lacks hormone receptor expression: progesterone receptor (PR) and estrogen (ER), and human epidermal growth factor receptor 2 (HER2) (3,4). Due to those characteristics, the treatment of patients with TNBC tumors is more challenging as compared to breast cancer patients with tumors of the other subtypes (4).
Recent studies have shown the importance of long non-coding RNAs (lncRNAs) in the progression of a wide range of malignant tumors such as gastric (5), prostate (6), esophagic(7), amongothers, (8,9), includingbreastcancer(10,11). lncRNAs are transcribed without coding for any
product, and are comprised of more than 200 nucleotides (12). These transcripts can be classified by several criteria including biological role, subcellular or genomic location, size, biological process, biogenesis pathway, or gene regulation (13).
For instance, the lncRNA H19, expressed only in the mother-inherited chromosome, is detected both in the nucleus and cytosol and is related to several biological processes such as inflammation, apoptosis, and angiogenesis (14). In breast cancer, H19 plays an important role in cellular proliferation, metastasis, chemoresistance, and stem cell maintenance (15). HOTAIR is another lncRNA, which is an important competitor with BRCA for the interaction with EZH2, which directly influences gene expression (16).
ADAMTS9-AS2 – ADAM metallopeptidase with thrombospondin type 1 motif, 9 antisense
RNA 2 – has an important role in tumoral progression and sensibility to chemotherapeutics (17). The
increase in ADAMTS9-AS2 expression and inhibition of miR-27a-3p in clear cell renal cell carcinoma (ccRCC) decreased cell proliferation and significantly reduced chemoresistance through regulation of FOXO1 (17). The knockdown of ADAMTS9-AS2 in squamous cell carcinoma of the tongue resulted in inhibition of cell migration and invasion and reverted the epithelial-mesenchymal transition process (EMT) induced by TGF-β1. Also, the study of Liet al. (2019) demonstrated that ADAMTS9-AS2 shares with EZH2 a binding site to miR-600, corroborating with the findings of
SONGet al. (2019) (17,18).
In the present study, we aimed to analyze the expression signatures of TNBCs from a microarray dataset, deposited in the GEO database, using the tool GEO2R. Based on the obtained results, we sought to investigate and validate the information available in international databases.
2 MATERIALS AND METHODS
2.1 GEO DATABASE ANALYSIS
The dataset of the microarray GSE76250 (19) was accessed through the platform GEO (https://www.ncbi.nlm.nih.gov/geo/) and analyzed using the online tool GEO2R (ncbi.nlm.nih.gov/geo/geo2r). The conversion of the IDs probes to the official name of each gene was performed by the BioGPS tool(http://biogps.org/#goto=searchresult). Genes were selected for further analysis according to the fold change ≥2.0 and p adjusted value by Benjamini& Hochberg (False Discovery rate) < 0.01 criteria.
2.2 UCSC
Long non-coding RNA conserved among human and other species and their chromosome localization were analyzed in UCSC genome browser portal (genome.ucsc.edu/).
2.3 EXPRESSION ANALYSIS
The tissues expression levels ware accessed by GTEx Project (gtexportal.org/) (20). We accessed the online database Breast Cancer Gene-Expression Miner v4.4 (bc-GenExMiner v4.4) (bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php), which is defined as a tool for statistical mining of transcriptomic data of breast cancer properly annotated. The databank contains microarray e RNA-seq information. Herein, we evaluated only RNA-seq-related data.
2.4 SURVIVAL ANALYSIS
We analyzed the chosen lncRNA and their association with overall survival (OS) and relapse-free survival (RFS) in the KM Plotter online database (kmplot.com/analysis/). Validated probes were chosen according to the automatic best cut off selection criteria. For the survival curves, patients were stratified by the lncRNAs expression according to intrinsic subtypes: all, basal, luminal A, luminal B, and HER2+.
2.5 TANRIC DATABASE
Gene expression data of 942 breast cancer patients were downloaded using the TANRIC platform (www.tanric.org/), of which 105 consisted of adjacent tissue samples and 837 were from tumor tissue. These data were cross-referenced with information available from TCGA (www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and UCSC Xena (xena.ucsc.edu / welcome-to-ucsc-xena /) to obtain clinicopathological data such as age, status survival, tumor stage, hormone receptors and human epidermal growth factor receptor type 2 (HER2). Cases of breast cancer or adjacent male breast tissue were excluded from this study.
2.6 CBIOPORTAL DATA
Expression data from 1.108 patients containing information about messenger RNAs (mRNAs) were downloaded on cBioPortal database (https://cbioportal.org). This information was cross-referenced with sample IDs from the TANRIC platform to pair samples for correlation analysis.
2.7 STATISTICAL ANALYSIS
The statistical analysis was performed using SPSS (Statistical Package for Social Sciences) version 25 and GraphPad Prism v. 7 (California, USA). The results were considered statistically significant when the p-value was less than 0.05 or, whenever necessary, according to p-adjusted values.
3 RESULTS
3.1 ADAMTS9-AS2 WAS IDENTIFIED IN MICROARRAY DATASET AND SHOWED DIFFERENT EXPRESSION IN MULTIPLE TISSUES
The GSE76250 aimed to determine the transcriptome of 165 triple-negative tumors (TNBC) and 33 paired normal breast tissue samples, using cDNA microarrays. Here, using the tool GEO2R, we compared TNBC samples with normal adjacent tissue and obtained a total of 70.523 probes, of which 19.238 showed an adjusted p-value less than 0.01 (Figure 1A). A probe representing the gene ADAMTS9-AS2 was identified at a high statistically significant level (5.43E-31; Supply. Table 1), and caught our interest for further analysis.
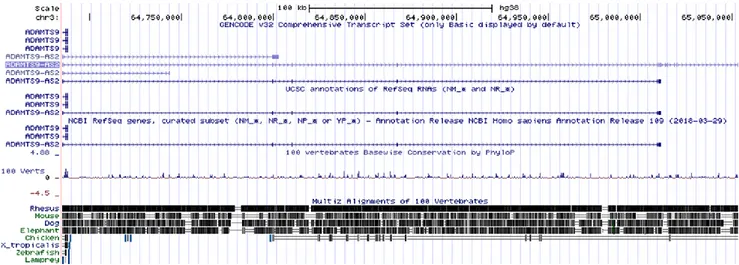
ADAMTS9-AS2 is a lncRNA, which maps on chromosome 3p14.1 with a 6 exon count. ADAMTS9-AS2 was not conserved among multiple species, with high similarity to rhesus (Figure 1B). The closest gene to ADAMTS9-AS2 is ADAMTS9, which is an antisense sequence and encodes a member of the large family of ADAMTS (A disintegrin and metalloproteinase with thrombospondin motifs 9) that are involved in tissue morphogenesis, pathophysiological remodeling, inflammation and vascular biology protein family (21). Analyzing data from GTEx project (20), ADAMTS9-AS2 was expressed in a diversity of tissues, including breast (Figure 1C).
Figure 1. A) Volcano Plot originated from the probes identified in the GSE76250, showing that lncRNA ADAMTS9-AS2 has the highest adjusted
p-value; B) Location of ADAMTS9-AS2 in human genome. Conservation in multiple species; C) Different tissue expression of ADAMTS9-AS2 shown by GTEx data.
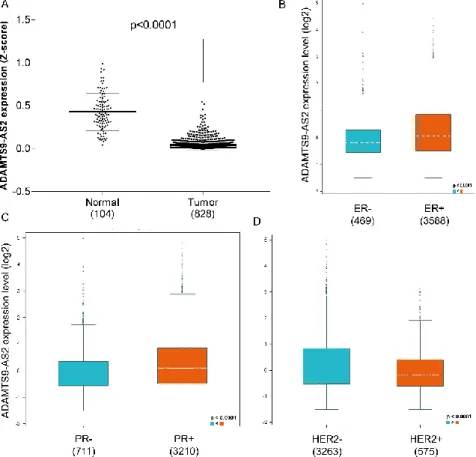
Figure 2. Expression of ADAMTS9-AS2. A) ADAMTS9-AS2 expression in normal and breast
tumor tissues provide by TANRIC data analysis. B) Expression of ADAMTS9-AS2 in function of the B) estrogen receptor and C) progesterone receptor; D) Expression of ADAMTS9
3.2 ADAMTS9-AS2 expression analysis
The expression of ADAMTS9-AS2 was evaluated by TCGA data downloaded and the online on bc-GenExMiner database. A significant difference was observed between adjacent and breast cancer tissue (p<0.0001; Figure 2A). We found a significant difference when evaluating the hormone receptors: ER and PR (Figure 2B-C), and an inverse relationship regarding expression was observed for HER2+ (Figure 2D).
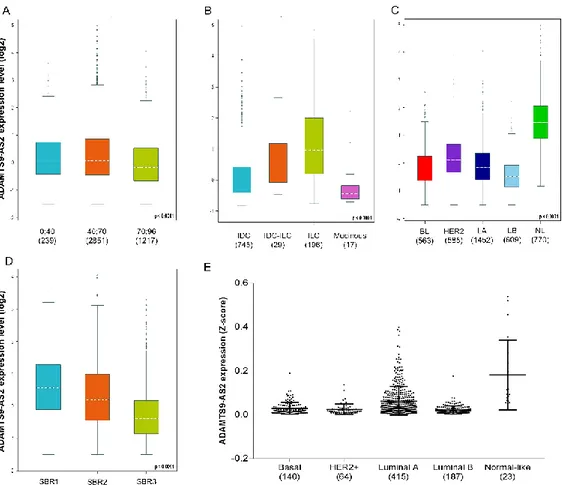
We also observed lower ADAMTS9-AS2 expression in older patients (Figure 3A). Regarding carcinoma type, IDC and mucinous showed the lowest transcript expression (Figure 3B). In regard to PAM50 classification, the basal and luminal B breast cancer subtypes exhibited low ADAMTS9-AS2 expression (Figure 3C). The Scarff-Bloom-Richardson (SBR) grade 3 breast tumors showed low ADAMTS9-AS2 expression (Figure 3D). The downloaded TCGA samples, which consist of 828 tumor breast tissues, were analyzed (Figure 3E) and similar result to bc-GenExMiner was observed, ensuring the reproducibility of our results.
Figure 3. Expression of ADAMTS9-AS2. A) Expression of ADAMTS9-AS2 in function of age of
breast cancer patients; B) Expression of ADAMTS9-AS2 in function of histological type; C) Expression of AS2 in function of PAM50 classification; D) Expression of ADAMTS9-AS2 in function of Scarff-Bloom-Richardson (SBR) grade classification; E) Expression of ADAMTS9-AS2 according to PAM50 provide by TANRIC data analysis. IDC: Invasive Ductal Carcinoma; ILC: Invasive Lobular Carcinoma; BL: Basal-like; HER2: human epidermal growth factor receptor 2-positive; LA: Luminal A; LB: Luminal B; NL: Normal-like. *** p-value less than <0.0001. Data obtained from Gene-Expression Miner v4.4 (http://bcgenex.centregauducheau.fr/BC-GEM/GEM-requete.php).
3.3 Expression of ADAMTS9-AS2 transcripts in basal and TNBC
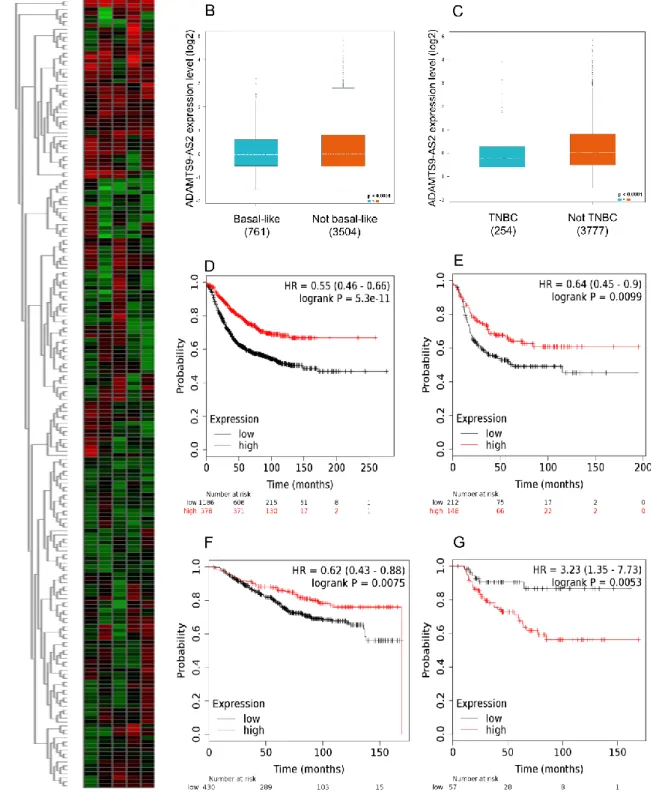
A heat map provided byR2: Genomics Analysis and Visualization Platform (r2.amc.nl/) used only with TNBC patients showed heterogeneous expression of ADAMTS9-AS2, suggesting different roles according to the TNBC subtypes (Figure 4A) (22,23). The bc-GenExMiner identified a lower expression of ADAMTS9-AS2 in Basal-like (Figure 4B) and TNBC (Figure 4C) subtypes. The relapse-free survival analysis identified a significant association between low expression of ADAMTS9-AS2 and worst prognosis, considering all breast cancer subtypes (Figure 4D). The same profile was observed when stratifying by the basal subtype (Figure 4E). The global survival showed a profile similar to the one found in RFS, however the basal subtype displays an inversion of the curve, in which high expression related to worst survival (Figure 4F-G).
Figure 4. Basal and TNBC expression and Kaplan-Meier of breast cancer patients stratified, according to ADAMTS9-AS2 expression. A) Heat Map with TNBC patients ADAMTS9-ADAMTS9-AS2 expression; B) Expression of ADAMTS9-ADAMTS9-AS2 in basal or not basal patients GenExMiner); C) Expression of ADAMTS9-AS2 in TNBC or not TNBC patients (bc-GenExMiner); Relapse-free survival for D) all patients and E) patients with basal breast tumores; Overall survival for F) all patients and G) patients with basal breast tumores. Survival analysis was conducted on the KM Plotter database (www.kmplot.com).
3.4 CO-EXPRESSION ANALYSIS
Co-expression of ADAMTS9-AS2 was conducted on TANRIC database. The co-expression profile of ADAMTS9-AS2 was identified with a large cluster composed of 1.155 genes, and RHOJ (ras homolog family member J) showed a high correlation (Figure 5A). Data mining in UCSC genome cancer, cBioPortal, and TANRIC databases confirmed the positive correlation between ADAMTS9-AS2 and RHOJ expression (Figure 5B-D).
Figure 5. Pearson correlation. A) The ADAMTS9-AS2 correlation was conducted on TANRIC database, which return 1155 different genes with a significant p-values; B) Heat map of ADAMTS9-AS2 and RHOJ by PAM50 classification. Data provide by UCSC genome cancer portal; Correlation analyze between ADAMTS9-AS2 and RHOJ on C) bc-GenExMiner and D) paired samples using TANRIC and cBioPortal data expression.
3.5 ADAMTS9-AS2 and clinical-pathological data associations
To validate the previous findings, we downloaded clinical-pathological data of patients with breast cancer, and data containing expression values of ADAMTS9-AS2. We utilized the expression median and categorized as high or low if above or below the found median, respectively. Significant associations were found in relation to age (p<0.0001), race (p=0.04), menopausal status (p=0.0003), PAM50 classification (p<0.0001), estrogen receptor (0.003), progesterone receptor (0.001), HER2 (0.0004) and mRNA RHOJ (<0.0001) (Tabel 1), previously identified in the TANRIC platform was positively correlated to ADAMTS9-AS2 (Figure 5).
ADAMTS9-AS2 expression was significantly lower in patients with tumor breast cancer, compared to the neighbor tissue (Figure 5A), and low expression was identified in basal tumors
(Figure 5B). We conducted a correlation analysis with paired samples containing expression data of ADAMTS9-AS2 and RHOJ, and found a positive correlation (r=0.725, p<0.0001; Figure 5C).
Table 1. Associations of ADAMTS9-AS2 expression and clinical-pathological data of patients with breast
cancer from TCGA database.
Parameters High Low
Total p-value n % N % Age <50 156 61.2 99 38,8 255 <0.0001*** >50 258 44.9 316 55,1 574 Race Category
American, Indian or Alaska native 0 0,0 1 100,0 1
0.0434*
Asian 24 45.3 29 54,7 53
Black or African american 25 39.7 38 60,3 63
White 341 54.4 286 45,6 627 Menopause Status Post 239 44.8 294 55,2 533 0.0003** Pre 114 60,0 76 40,0 190 Metastasis No 371 50.1 369 49,9 740 0.1350 Yes 5 31.3 11 68,8 16 Stage (TNM) I/II 297 50,0 297 50,0 594 0.8157 III/IV 110 50.9 106 49,1 216 PAM50 classification Basal 57 40.7 83 59,3 140 <0.0001 HER2+ 18 28.1 46 71,9 64 Luminal A 270 65.1 145 34,9 415 Luminal B 47 25.1 140 74,9 187 Normal-like 22 95.7 1 4,3 23 Radiation therapy No 163 49.8 164 50,2 327 0.6659 Yes 214 51.4 202 48,6 416 ER Status By IHC Negative 75 41,0 108 59,0 183 0.0031** Positive 325 53.5 283 46,5 608 PR status by IHC Negative 108 42,0 149 58,0 257 0.0009** Positive 290 54.6 241 45,4 531
HER2 status by IHC
Negative 242 55.1 197 44,9 439 0.0004** Positive 48 37.5 80 62,5 128 RHOJ High 328 79.2 86 20,8 414 <0.0001 Low 85 20.5 329 79,5 414 OS status Alive 356 50.7 346 49,3 702 0.2956 Dead 58 45.7 69 54,3 127
ER: estrogen receptor; PR: progesterone receptor; HER2: human epidermal growth factor receptor 2; RHOJ: ras homolog family member J; OS: Overall survival.
4 DISCUSSION/CONCLUSION
Among several dysregulated lncRNAs, ADAMTS9-AS2 was identified to be the most down-regulated lncRNA in several tumor types, including breast cancers (24,25). The role played by ADAMTS9‐AS2 has been demonstrated in a wide range of functional networks, including cell migration, proliferation and epithelial-mesenchymal transition (EMT) across different types of cancer (17,18). Recently, Deva Magendhra Rao et al. (2019) identified ADAMTS9‐AS2 expression as frequently down-regulated in breast tumors and provided evidence that promoter region hypermethylation is the main event associated ADAMTS9‐AS2 reduced expression in breast cancer (Deva Magendhra Rao et al., 2019). In addition, the downregulation of ADAMTS9-AS2 has been associated with chemoresistance against some carcinomas (17). For instance, SHI; LU and WANG (2019) reported an association between this lncRNA and tamoxifen resistance by the activation of a microRNA in the breast cancer cell line MCF-7. In agreement with these previous reports, our findings provide additional evidence of the significant role of the lncRNA ADAMTS9-AS2 in breast cancer, and its association with progesterone and estrogen receptors.
Here, we also found that ADAMTS9-AS2 was associated with the worst prognostics for all breast cancer subtypes and, interestingly, significantly less expressed in older patients. Whether the reduced levels in the elderly maybe be due to a progressive reduction in its expression because of natural aging or may be caused by a sudden and random disruption is an important question that needs further clarification. In addition, this transcript was downregulated in TNBC patients, and low levels were associated with a worse prognosis. The overall survival (OS) shows the opposite since high levels of ADAMTS9-AS2 were associated with a more aggressive disease. This may happen because of the OS estimate considers all causes of death. In other words, a robust analysis with only deaths due to breast cancer is necessary to understand the role of ADAMTS9-AS2 levels and its real impacts on the survival fo TNBC patients.
It is important to note that the expression of ADAMTS9-AS2 seems to behave differently among TNBC patients, which can be explained by the variety of intrinsic subtypes found by molecular classification: Basal-like 1 (BL1), Basal-like 2 (BL2),immunomodulatory (IM), mesenchymal (M), mesenchymal stem-like (MSL), and luminal androgen receptor (LAR) subtype; and an unstable (UNS) subtype (22,23).
Furthermore, we successfully found a strong correlation between ADAMTS9-AS2 and RHOJ expression levels. This latter gene codes for a GTP-binding protein, which has been implicated in the regulation of melanoma chemoresistance (27) and breast cancer cell migration through interaction with the EVI1 oncogene as part of the GPR signaling pathway (28). RHOJ is also linked to other functional networks such as the cytoskeleton, adhesion, infection, and Ras signaling pathways, as
shown in a recent integrated bioinformatics analysis that assessed RHOJ’s expression levels in NonSmall Cell Lung Cancer (29). Since ADAMTA-AS2 is highly correlated to RHOJ, it is possible that this lncRNA might also play a germane role in cell migration as well as in other functional networks and molecular mechanisms.
In this context, unraveling the molecular mechanisms by which the expression of ADAMTS9-S2 is regulated and how it might act on cancer progression becomes necessary and should be accomplished in further in vitro experiments. However, it should be noted that there is high heterogeneity in breast cancer tumors across individuals as well as across different tumor progression stages (2,30). Hence, these tumor and patient-specific aspects should be taken into account when conducting these experiments in order to provide more precise and accurate conclusions. These future investigations might provide new information to the design of new and more effective therapies for breast cancer patients, especially the ones with harsher prognostics such as triple-negative individuals in older ages.
In the evaluated dataset GSE76250, low expression of ADAMTS9-AS2 was significantly lower in triple-negative breast tumors compared to adjacent mammary tissues. Taking together with the available reports in the literature on other anatomical sites and in breast cancer, our findings indicate ADAMTS9-AS2 might be a potential biomarker for TNBC patients.
5 SUPPLEMENTARY MATERIAL
Table S1. The top 100 probes classified by adjusted p-value.
ACKNOWLEDGMENT
This work was supported by CNPq (National Council for Scientific and Technological Development) and CAPES (Coordination for Higher Education Personnel Improvement).
AUTHOR CONTRIBUTIONS
Daniel Rodrigues de Bastos, Jean Michel Rocha Sampaio Leite and Mércia Patrícia Ferreira Conceição performed the analyses and interpretation of data. Maria Aparecida Nagai, Antônio Marcio Teodoro Cordeiro Silva and Cesar Augusto Sam Tiago Vilanova-Costa supervised the findings of this work and revised it critically. All authors discussed the results and contributed with drafting of the final manuscript.
REFERENCES
1. Werutsky G, Nunes P, Barrios C. Locally advanced breast cancer in Brazil: Current status and future perspectives. Ecancermedicalscience. 2019 Jan 22;13.
2. McDonald KA, Kawaguchi T, Qi Q, Peng X, Asaoka M, Young J, et al. Tumor Heterogeneity Correlates with Less Immune Response and Worse Survival in Breast Cancer Patients. Ann Surg Oncol. 2019 Jul 15;26(7):2191–9.
3. Aysola K, Desai A, Welch C, Xu J, Qin Y, Reddy V, et al. Triple Negative Breast Cancer-An Overview. Hered Genet. 2013;2013(2).
4. Collignon J, Lousberg L, Schroeder H, Jerusalem G. Triple-negative breast cancer: treatment challenges and solutions. 2016 [cited 2020 Jan 13]; Available from: http://dx.doi.org/10.2147/BCTT.S69488
5. Qi M, Yu B, Yu H, Li F. Integrated analysis of a ceRNA network reveals potential prognostic lncRNAs in gastric cancer. Cancer Med [Internet]. 2020 Jan 10 [cited 2020 Jan 13];cam4.2760. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/cam4.2760
6. Shi X, Zhang W, Nian X, Lu X, Li Y, Liu F, et al. The previously uncharacterized lncRNA APP promotes prostate cancer progression by acting as a competing endogenous RNA. Int J Cancer. 2019;
7. Wang L, Meng D, Wang Y, Hu J. Long non-coding RNA LINC01296 promotes esophageal squamous cell carcinoma cell proliferation and invasion by epigenetic suppression of KLF2. Am J Cancer Res [Internet]. 2018 [cited 2020 Jan 13];8(10):2020–9. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30416853
8. Yang Y, Chen Q, Piao H, Wang B, Zhu G, Chen E, et al. HNRNPAB‐regulated lncRNA‐ ELF209 inhibits the malignancy of hepatocellular carcinoma. Int J Cancer [Internet]. 2020 Jan 17 [cited 2020 Jan 13];146(1):169–80. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/ijc.32409
9. Liu L, Zhan Y, Huang Y, Huang L. LncRNA FGD5‐AS1 can be predicted as therapeutic target in oral cancer. J Oral Pathol Med [Internet]. 2020 Jan 3 [cited 2020 Jan 13];jop.12989. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/jop.12989
10. Cui Y, Lu C, Zhang Z, Mao A, Feng L, Fu L, et al. A Long Non-coding RNA Lnc712 Regulates Breast Cancer Cell Proliferation. Int J Biol Sci [Internet]. 2020 [cited 2020 Jan 13];16(1):162–71. Available from: http://www.ijbs.com/v16p0162.htm
11. Giro-Perafita A, Luo L, Khodadadi-Jamayran A, Thompson M, Akgol Oksuz B, Tsirigos A, et al. LncRNA RP11-19E11 is an E2F1 target required for proliferation and survival of basal breast
cancer. npj Breast Cancer [Internet]. 2020 Dec 6 [cited 2020 Jan 13];6(1):1. Available from: http://www.nature.com/articles/s41523-019-0144-4
12. Dahariya S, Paddibhatla I, Kumar S, Raghuwanshi S, Pallepati A, Gutti RK. Long non-coding RNA: Classification, biogenesis and functions in blood cells. Vol. 112, Molecular Immunology. Elsevier Ltd; 2019. p. 82–92.
13. Jarroux J, Morillon A, Pinskaya M. History, discovery, and classification of lncRNAs. In: Advances in Experimental Medicine and Biology. Springer New York LLC; 2017. p. 1–46.
14. Wang J, Sun J, Yang F. The role of long non-coding RNA H19 in breast cancer (Review). Oncol Lett. 2020;19(1):7–16.
15. Wang J, Xie S, Yang J, Xiong H, Jia Y, Zhou Y, et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J Hematol Oncol [Internet]. 2019 Dec 24 [cited
2020 Jan 13];12(1):81. Available from:
https://jhoonline.biomedcentral.com/articles/10.1186/s13045-019-0747-0
16. Pawłowska E, Szczepanska J, Blasiak J. The long noncoding RNA HOTAIR in breast cancer: Does autophagy play a role? Int J Mol Sci [Internet]. 2017 [cited 2020 Jan 13];18(11). Available from: http://www.genecards.org/cgi-bin/carddisp.pl?gene=HOTAIR;
17. Song EL, Xing L, Wang L, Song WT, Li D Bin, Wang Y, et al. LncRNA ADAMTS9-AS2 inhibits cell proliferation and decreases chemoresistance in clear cell renal cell carcinoma via the miR-27a-3p/FOXO1 axis. Aging (Albany NY). 2019 Aug 15;11(15):5705–25.
18. Li Y, Wan Q, Wang W, Mai L, Sha L, Mashrah M, et al. LncRNA ADAMTS9-AS2 promotes tongue squamous cell carcinoma proliferation, migration and EMT via the miR-600/EZH2 axis. Biomed Pharmacother. 2019 Apr 1;112.
19. Jiang YZ, Liu YR, Xu XE, Jin X, Hu X, Yu K Da, et al. Transcriptome Analysis of Triple-Negative Breast Cancer Reveals an Integrated mRNA-lncRNA Signature with Predictive and Prognostic Value. Vol. 76, Cancer Research. American Association for Cancer Research Inc.; 2016. p. 2105–14.
20. GTEx Project. GTEx portal. GTEx Analysis Release V6p (dbGaP Accession phs000424.v6.p1). 2017.
21. Shao B, Feng Y, Zhang H, Yu F, Li Q, Tan C, et al. The 3p14.2 tumour suppressor ADAMTS9 is inactivated by promoter CpG methylation and inhibits tumour cell growth in breast cancer. J Cell Mol Med. 2018 Feb 1;22(2):1257–71.
22. Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011 Jul;121(7):2750–67.
23. Santonja A, Sánchez-Muñoz A, Lluch A, Chica-Parrado MR, Albanell J, Chacón JI, et al. Triple negative breast cancer subtypes and pathologic complete response rate to neoadjuvant chemotherapy. Oncotarget. 2018 May 29;9(41):26406–16.
24. Deva Magendhra Rao AK, Patel K, Korivi Jyothiraj S, Meenakumari B, Sundersingh S, Sridevi V, et al. Identification of lncRNAs associated with early-stage breast cancer and their prognostic implications. Mol Oncol. 2019 Jun 1;13(6):1342–55.
25. Yao J, Zhou B, Zhang J, Geng P, Liu K, Zhu Y, et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumor Biol [Internet]. 2014 Aug 15;35(8):7935–44. Available from: http://link.springer.com/10.1007/s13277-014-1949-2
26. Shi YF, Lu H, Wang HB. Downregulated lncRNA ADAMTS9-AS2 in breast cancer enhances tamoxifen resistance by activating microRNA-130a-5p. Eur Rev Med Pharmacol Sci. 2019;23(4):1563–73.
27. Ho H, Aruri J, Kapadia R, Mehr H, White MA, Ganesan AK. RhoJ regulates melanoma chemoresistance by suppressing pathways that sense DNA damage. Cancer Res [Internet]. 2012 Nov 1;72(21):5516–28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22971344
28. Wang H, Schaefer T, Konantz M, Braun M, Varga Z, Paczulla AM, et al. Prominent Oncogenic Roles of EVI1 in Breast Carcinoma. Cancer Res [Internet]. 2017;77(8):2148–60. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28209621
29. Zeng T, Chen C, Yang P, Zuo W, Liu X, Zhang Y. A Protective Role for RHOJ in NonSmall Cell Lung Cancer Based on Integrated Bioinformatics Analysis. J Comput Biol [Internet]. 2019 Oct 23; Available from: http://www.ncbi.nlm.nih.gov/pubmed/31638412
30. Liu YR, Jiang YZ, Xu XE, Hu X, Yu K Da, Shao ZM. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin Cancer Res. 2016 Apr 1;22(7):1653–62.