staphylococcus aureus clones in Latin America:
implications for clinical practice in the region
aBstract
Methicillin-resistant Staphylococcus aureus (MRSA) clones belonging to the Brazilian, Pediatric, Cordobes/Chilean and New York/Japan clonal complexes are widely distributed across Latin Ameri-ca, although their individual distribution patterns and resistance to antimicrobial drugs are constantly changing. Furthermore, clones with increased virulence are beginning to appear more frequently both in hospital and community settings, and there is evidence that virulence factors can be transferred be-tween hospital- and community-associated clones through recombination. These changing patterns have significant implications for clinical practice in the region. Most importantly, clinicians need to be aware of the changing antimicrobial resistance profile of circulating MRSA clones in their region in order to choose the most appropriate empiric antimicrobial therapy. Thus, regional molecular epidemiology pro-grams are required across the region to provide accurate identification and characterization of circulating MRSA clones.
Keywords:MRSA,clones,molecularepidemiology,LatinAmerica.
introDuction
Methicillin-resistant Staphylococcus aureus
(MRSA) poses a major threat to public health worldwide,duetotherapidspreadanddiversifi-cationofpandemicMRSAcloneswithincreasing virulence and antimicrobial resistance. In Latin America,MRSAisaleadingcauseofnosocomial infections,andtheprevalenceofMRSAincom-munity-acquiredinfectionsisgrowing.1
Although relatively few studies have ad-dressed the molecular epidemiology of MRSA clonesacrossLatinAmerica,itisclearthatseveral clonescirculateintheregionandthatthesediffer intheirvirulence,antimicrobialresistanceprofile and geographical distribution.1,2
Characteriza-tion of these clones is important if appropriate localtreatmentstrategiesaretobedeveloped.For example,athoroughknowledgeofclonescircu-latingwithinaregionmaybeusedtoassessthe relationshipbetweenclonaltypes,diseasesymp-toms, antibiotic choice and clinical outcomes. Furthermore,understandingwhyspecificclones
predominateindifferentregionsofLatinAmer-ica is an important and necessary step towards developingthemosteffectivestrategiesforcon-trollingthespreadofMRSAintheregion.
Here, we summarize current understand-ingofthespreadofpandemicMRSAclonesand highlightthedistributionofmajorclonesacross LatinAmerica,bothinhospitalsandinthecom-munity. Specific virulence factors and bacterial resistancepatternsarehighlighted,andtheirim-pactonclinicaloutcomeisdiscussed.
evolution of Mrsa clones
Evolution of bacterial clones
Bacterialclonesaregeneticallyidenticalcellsde-scended from a single common ancestor. Over time,membersofasingleclonemaydifferenti-atethroughpointmutations,recombination,and theacquisitionordeletionofmobilegeneticel-ements. This differentiation provides additional
meansfortheacquisitionofpathogeniccharac-Authors
EduardoRodríguez-Noriega1
CarlosSeas2
onbehalfoftheLatin AmericanWorking GrouponGram PositiveResistance.
1HospitalCivil
deGuadalajara, Universidadde Guadalajara,Jalisco, Mexico.
2Universidad
PeruanaCayetano Heredia,Lima,Peru.
Correspondence to:
DrEduardo Rodríguez-Noriega HospitalCivilde Guadalajara FrayAntonioAlcalde InstitutodePatología Infecciosay Experimental CentroUniversitario CienciasdelaSalud Universidadde Guadalajara Jalisco,México Hospital308, ColoniaElRetiro C.P.44280, Guadalajara Jalisco,México Phone:+52-33-3614-5568
Fax:+52-33-3685-0501
teristics,suchasantibioticresistance.Thus,geneticvariation givesrisetoextensivegenomicandphenotypicdiversity.
Emergence of antibiotic resistant S. aureus clones Clones ofS. aureus have a history of antibiotic resistance thatbeganwithin4yearsoftheintroductionofpenicillin intoclinicalpractice,3andby1944,clonesofS. aureus
resist-anttopenicillinhadbeenisolated.Inthesubsequentyears,
S. aureusbecameresistanttoallofthenaturalpenicillins. MRSAwasfirstreportedintheearly1960s,shortlyaf-tertheintroductionofmethicillin.4
EarlyMRSAcloneshad similar genetic properties to the methicillin-susceptibleS. aureus(MSSA)clonesthatwereepidemicinEurope.5MRSA
exhibitsresistancetomethicillinthroughapenicillin-bind-ingproteinencodedbythegenemecA,whichwasacquired bysuccessfulclonesofMSSAfromanunknownheterolo-goussource.ThemecA geneiscarriedbyamobilegenetic elementcalledthestaphylococcalcassettechromosomemec
(SCCmec).MultipleformsofSCCmechavearisenthrough thehorizontaltransferofmecAinindependentevents,and, todate,sevenmainformshavebeenidentified(I,II,III,IV, V, VI and VII).6 All types of SCCmec confer resistance to
β-lactam antibiotics, and SCCmec types II and III provide resistancetomultipleclassesofantibiotics.7
DuringtheevolutionofMRSAclones,independentex-cision of SCCmec is a common phenomenon, resulting in thelossofmethicillin-resistanceandthetransformationof aMRSAcloneintoaMSSAclone.Hence,clonesmayevolve fromMSSAintoMRSA,orfromMRSAintoMSSA,through theacquisitionandexcisionofSCCmec,respectively.8
Several molecular typing methods are used routinely to characterizeMRSAclones,includingpulsed-fieldgelelectro-phoresis (PFGE), multilocus sequence typing (MLST) and SCCmectyping.9Thesemethodshavehelpedresearchersto
mapthespreadandevolutionarypathofMRSAclones.7,8
MRSAhastraditionallybeenregardedasanosocomial pathogen,10 but more recently, MRSA infections have
ap-peared in community settings.11
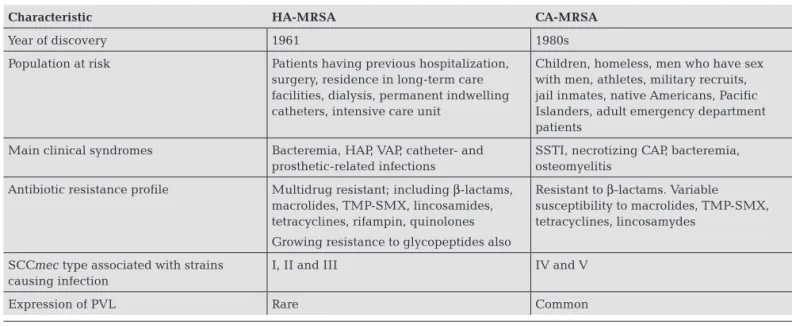
The clones typically re- sponsibleforhospital-andcommunity-acquiredMRSAin-fectionshavebeenclassifiedashealthcare-associatedMRSA (HA-MRSA) and community-associated MRSA (CA-MR-SA),respectively.12Theseclonescanbedistinguishedbased
on specific microbiologic and genetic characteristics, and oftenhavedifferentepidemiologic,clinicalandtherapeutic characteristics(Table1).10,11Occasionally,hospital-acquired
infectionsmaybederivedfromCA-MRSAstrains,andin- fectionsacquiredinthecommunitymaycarryhealthcare-associatedriskfactors.DefinitiveHA-MRSAandCA-MRSA designationsforindividualclones,therefore,relyonmicro- biologicandgeneticcharacterization,andtheterms‘health- care-acquired’and‘community-acquired’refertotheloca-tionatwhichtheinfectionwasacquired.12
International spread of MRSA clones
S. aureusclonesspreadquicklyaroundtheworld,13
dissemi-natingefficientlybetweencountries,withincountriesandin smallergeographicareas,andusuallywithconcomitantevo-lutionfromamethicillin-sensitivetoamethicillin-resistant phenotype.5,14 Most nosocomial MRSA infections
world-wide are derived from one of five major lineages known
table 1. common characteristics of infections caused by ha-Mrsa and ca-Mrsa
Characteristic HA-MrSA CA-MrSA
Year of discovery 1961 1980s
Population at risk Patients having previous hospitalization,
surgery, residence in long-term care facilities, dialysis, permanent indwelling catheters, intensive care unit
Children, homeless, men who have sex with men, athletes, military recruits, jail inmates, native Americans, Pacific Islanders, adult emergency department patients
Main clinical syndromes Bacteremia, HAP, VAP, catheter- and
prosthetic-related infections
SSTI, necrotizing CAP, bacteremia, osteomyelitis
Antibiotic resistance profile Multidrug resistant; including β-lactams,
macrolides, TMP-SMX, lincosamides, tetracyclines, rifampin, quinolones
Growing resistance to glycopeptides also
Resistant to β-lactams. Variable susceptibility to macrolides, TMP-SMX, tetracyclines, lincosamydes
SCCmec type associated with strains causing infection
I, II and III IV and V
Expression of PVL Rare Common
asclonalcomplexes(CC):5,8,22,30and45.7,15,16Between
1994and2000,surveillancedatacollectedbytheCEM/NET initiativeidentifiedfivepredominantpandemicclones(Bra-zilian, Iberian, Hungarian, pediatric and New York/Japan [NYJ])withintheseclonalcomplexes,andtheseaccounted fornearly70%ofMRSAisolatesworldwide.17
The intercontinental spread of MRSA derived from a singleancestralMRSAclonewasobservedintheUSA,UK, Denmark,Switzerland,EgyptandUgandaintheearly1990s. In the late 1990s, further spread was noted when MRSA clones previously isolated from Australia, US and Ireland, werefoundtohavesimilaritieswithotherMRSAclonescol-lectedaroundtheworld.13,18
Withincountries,MRSAcloneshavedisseminatedrap-idlyandefficiently.Diversificationanddisplacementofthese clonesissimilarlyrapid.Overa9-yearperiodinSpain,the common NYJ MRSA clone was displaced by the Brazilian MRSAclone,whichwasthendisplacedbythenow-predom-inant EMRSA-16 clone.19 Similarly, in Belgium, epidemic
MRSAclonesbelongingtoclonalcomplexes5,8,22,30and 45diversifiedrapidly,andby2001,theseclonalcomplexes showedbroadgeographicdistributioninBelgianhospitals, incontrasttoearliersurveyresults.15
InPortugal,thepre-dominantBrazilianMRSAclonewasreplacedinhospitals bytwoearlierMRSAclonesovera16-yearperiod.16
IntheUSA,HA-MRSAclonesarerapidlybeingreplaced byCA-MRSAclones.Theextentofthisreplacementvaries betweenareas,butinoneChicagohospitaltheproportion of community-associated isolates increased from 24% to 49%over3years.20
Mrsa clones in latin aMerica
Since the first report in 1994 of an autochthonous MRSA cloneoriginatinginBrazil,21knownastheBrazilianclone,
four additional clones circulating in Latin America have beendescribed;theCordobes,thePediatric,theChileanand theNYJclones.22-24
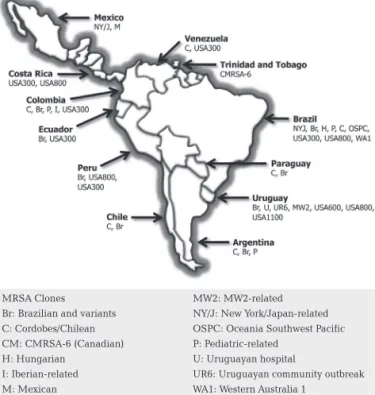
Theclosely-relatedCordobesandChil- eanclonesarenowconsideredtobeasingleclone(Cordo-bes/Chilean).Alloftheseclonescirculateextensivelyinthe region(Figure123-36),andthereisevidencesupportingthe
appearance of genetic variants. Several minor clones have also been identified, although these currently occur in re-strictedgeographicareas(Figure123-36).37
TheBrazilianclone(ST239-SCCmectypeIII)hasbeen isolated throughout Brazil and has extended into several othercountries,includingArgentina,Chile,Colombia,Ec-uador, Paraguay, Peru and Uruguay. Several genetic vari-antshavebeenidentified,andtogetherthesearetermedthe Brazilian epidemic clonal complex (BECC).26 The
patho- genicityoftheBrazilianclonederivesfromseveralproper- ties,whicharepresenttodifferentdegreesindifferentvari-ants:multidrugresistance,includingresistancetoβ-lactams, chloramphenicol, quinolones, lincosamides,
erythromy-cin, aminoglycosides and trimethoprim-sulfamethoxazole (TMP-SMX); resistance to mupirocin;38 production of a
protectivebiofilm;26abilitytoadhereto,andinvade,airway
epithelialcells;26
andproductionoftoxins,suchasentero-toxinsandPanton-Valentineleukocidin(PVL).34
TheCordobesandChileanclones(ST5-SCCmectypeI) were identified separately in isolates fromArgentina23 and
Chile,22andwerelaterconsideredtobevariantsofthesame
clone.InArgentina,theCordobes/Chileanclonehasrapidly replacedtheclosely-relatedBrazilianclone,anditnowpre-dominates in several Latin American countries, including Argentina, Chile, Paraguay and Colombia,27,32,35 where it is
associatedwithnosocomialoutbreaks.TheCordobes/Chil-ean clone shows a multidrug resistant phenotype, includ-ing resistance to erythromycin, although variants are still susceptibletoglycopeptides,linezolid,TMP-SMX,rifampin andtetracyclines.
Thepediatric(USA800;ST5-SCCmectypeIVorSCCmec
type VI) and NYJ (USA100; ST5-SCCmec type II) MRSA cloneshavealsospreadsuccessfullythroughLatinAmerica. Variantsofthepediatricclone,withheterogeneousandlow- levelresistancetomethicillinandresistancetob-lactaman-timicrobials,havecausedinfectionsinBrazil,Argentinaand Colombia,23,30,39andhavedevelopedmultidrugresistancein
somehospitalsinLatinAmerica.30TheNYJclone,typically
resistanttoβ-lactams,erythromycin,clindamycinandcipro-floxacin,28,33hasbeendetectedinBrazilandhascompletely
displacedtheMexicancloneincertainMexicanhospitals.24
InotherLatinAmericancountries,fewdataareavailable todescribethespreadofindividualMRSAclonesfully.Ina
Figure 1: MRSA clones circulating in Latin America identified since 200023-36
MRSA Clones Br: Brazilian and variants C: Cordobes/Chilean CM: CMRSA-6 (Canadian) H: Hungarian
I: Iberian-related M: Mexican
MW2: MW2-related NY/J: New York/Japan-related OSPC: Oceania Southwest Pacific P: Pediatric-related
U: Uruguayan hospital
recentstudy,PVL-negativeST5-SCCmectypeIV(USA800) and ST8-SCCmec type IV (USA300) MRSA clones were identifiedinCostaRica,andaPVL-positiveST5-SCCmecIV clone(USA800)wasidentifiedinPeru.29
InTrinidadandTo-bago,theCanadianMRSAclone,CMRSA-6,wasobserved inseveralmajorhospitalsbetween2000and2001,possibly asaresultoftourism.25
The first CA-MRSA infections to be reported in Latin America occurred in Brazil in 2003,34 where isolates from
patientswithskinandsofttissueinfections(SSTIs)orseptic arthritis,werefoundtoharborSCCmectypeIVandPVL. InUruguay,alargeoutbreakofCA-MRSAinfectionsinjail inmates occurred,37 in which > 1000 patients were
docu-mented with SSTIs and severe forms of pneumonia. This outbreak was caused by a CA-MRSA clone harboring SC-CmectypeIVandPVL.CA-MRSAclonesbelongingtoCC5, CC8andCC30havenowbeenidentifiedinBrazil,Argen-tina,ColombiaandUruguay,andthesearerelatedtoclones previouslyobservedintheUSAandAustralia.Inaddition, community-acquiredinfectionsduetoMRSAhavebeenre-portedinPeru,40Venezuela41
andChile,42,43althoughsome
ofthesecaseswereinpeoplereturningfromcitiesinUru- guayorBrazilwhereahigherincidenceofcommunity-ac-quiredMRSAinfectionsoccurred.Inarecentstudyinfour South American countries (Colombia, Ecuador, Peru and Venezuela), Arias and colleagues found a new CA-MRSA USA300variantintheAndeanregion.36
clinical iMplications of Mrsa clones
Virulence and antibiotic resistance of MRSA clones
MRSA strainscauseavarietyofinfections,ranginginsever-ityfromcutaneousabscesses,tolife-threateningnecrotizing fasciitisandnecrotizingpneumonia.Theseverityofdisease isusuallydirectlyrelatedtotheproductionbyMRSAofspe-cificvirulencefactors,44
suchastoxinsorprotectivebiofilms, whereasthespreadofMRSAdependspartlyontheabilityof individualclonestoacquireresistancetoantibacterialagents.
Panton-Valentine Leukocidin Production
PVL,amajorvirulencefactorforMRSA,isasecretedtoxin thatcausestissuenecrosisanddamagetoimmunecells.The geneencodingPVL,pvl, encodestwosecretedsubunits,des-ignatedLukS-PVandLukF-PV,whichassembletogetherin themembranesofwhitebloodcells,monocytesandmac-rophagestoformporesthroughwhichcellcontentsleak.45
PVL-producingMRSAstrainshavebeenassociatedwithSS-TIsandasevereformofnecrotizingpneumonia.46,47InaUS
studyof422patientswithcommunity-acquiredSSTIs,59% ofisolateswereidentifiedasMRSA(mainlycloneUSA300), and 98% of these expressed PVL,48 consistent with other
studies.49ThepresenceofPVLhasnowbeendemonstrated
inbothadultandpediatricMRSAinfections.49,50
AcquisitionofpvlbyCA-MRSAclonesmarkedadramatic changeintheepidemiologyofMRSAinfections,bothinthe communityand,morerecently,inhospitals.51-53
Severalstud-ieshavereportedtheacquisitionofpvl byMRSAclonescircu-latinginLatinAmerica.34,54,55InRiodeJaneiro,forexample,
BECCisolateswereshowntobepositiveforpvl ,possiblyfol-lowing horizontal transfer from a reservoir of PVL-positive MSSAisolates.54
InArgentina,94%ofisolatesofthepredomi-nantCA-MRSAcloneST5werefoundtoharborpvl.35
Biofilm production by MRSA strains
S. aureusstrainsroutinelyproduceandbecomeencasedin biofilm,providingthemwithprotectionagainstbothhost defensesandantimicrobialdrugs.Biofilmproductionisme-diatedbytheicaoperon,andthispropertyispresentinmost MRSA and MSSA strains, although evidence suggests that certainMRSAcloneshaveanenhancedabilityinthisregard. In2005,thepredominantvariantoftheBECCMRSAclone wasreportedtobemoreeffectiveingeneratingbiofilmand in adhering to, and invading, airway epithelial cells, than MSSAorsporadicMRSAclones.26InarecentBrazilianstudy
ofMRSAisolatesfromhospital-acquiredandcommunity-acquiredMRSAinfections,all19MRSAstrains(14Brazilian clone;5NYJclone)werefoundtoproducebiofilm.56
Resistance in MRSA clones
MRSA,andinsomecountriesvancomycin-resistant entero-cocci ,arethemostfrequentantibiotic-resistantgram-posi-tive bacteria responsible for nosocomial infections and, in thecaseofMRSA,communityinfections.57
HA-MRSA strains are often multidrug resistant con-ferredbySCCmectypesIIandIII,58
andcanexpressresist- ancetofluoroquinolones,macrolides,aminoglycosides,tet-racyclinesandrifampin.MultidrugresistantMRSAstrains canalsodevelopreducedsusceptibilityorresistancetovan-comycinandteicoplanin,aswellastonewerantimicrobials suchasquinupristin-dalfopristin,linezolid,daptomycinand tigecycline,althoughtheseoccurrencesarelimitedtoafew isolatedcases.S. aureus withreducedsusceptibilitytovanco-mycin,orvancomycin-intermediateS. aureus (VISA),usu- allydevelopthislimitedsusceptibilitybychangingthethick-nessoftheircellwall.Consequently,vancomycinbecomes trappedintheouterwallandhasreducedaccesstoitstarget inthecytoplasmicmembrane. S. aureus canalsodevelopfull resistance to vancomycin (VRSA), which is thought to be linkedtotheacquisitionoftheresistancegenevanAfrom vancomycin-resistantenterococci.59,60
bymacrolides,andcanbedetectedfollowingerythromycin-mediatedinductionusingtheapproximationtest.61Studies
basedintheUSAreporttheoverallprevalenceofinducible clindamycin resistance inS. aureus isolates as 52% (50% ofMRSAand60%ofMSSAisolates),62althoughthismay
change,sinceclonalshifthasbeenshowntoaffectinducible clindamycinresistance.63
InLatinAmerica,MRSAclonesaregeneticallydiverse,but oftenshareincommongenesthatencodeformultidrugresist-ance to β-lactams, erythromycin, chloramphenicol and clin-damycin,andvariableresistancetorifampin,fluorquinolones andTMP-SMX.Threeclassesofantimicrobials,includingglyc-opeptides,oxazolidinonesandthenewtetracyclinederivative tigecycline,areuniformlyactiveagainsttheseclonalvariantsin theregion.TheBraziliancloneissometimesonlysensitiveto vancomycin, tigecycline, linezolid and daptomycin.Although therearereportsindicatingheteroresistancetovancomycinin theBrazilianclone,thereiscurrentlylittleevidencetosuggest thatVISAand/orVRSAareasignificantissueinLatinAmerica. However,itwillbeimportanttomonitorthesituationacross theregioninordertoidentifyanychangestothepatternofan- timicrobialresistanceasearlyaspossible.TheCordobes/Chil-eanandNYJclones,andinsomeplacesalsothepediatricclone, aremultidrugresistant.Usually,theCordobes/Chileancloneis susceptibletoglycopeptides,linezolid,TMP-SMX,minocycline andrifampin,whiletheNYJcloneissensitivetoglycopeptides, linezolid,TMP-SMX,rifampinandgentamicin.Carefuluseof antibioticsisanimportantstrategyforlimitingantimicrobial resistance. For example, high level resistance of the Brazilian clonetomupirocinhasbeenreportedinBrazilianhospitals,38
butrecentevidencesuggeststhatresistancecanbereducedby controllingmupirocinuse.64
MRSA clones in hospital and community settings
Isolates from nosocomial MRSA infections, typically col-lected>72hoursfollowinghospitaladmission,usually,but notalways,containMRSAclonesharboringSCCmectypeI, IIorIII(Table1).Theseclonesproducebacteremia,pneu-moniaorurinarytractinfectionsmorefrequentlythanthey causeSSTI.31,65NosocomialMRSAisusuallyreportedasan
adultpathogen,buthasalsobeenassociatedwithinfections inpediatricandneonatalintensivecareunits.24,66
Isolates from community-acquired MRSA infections, typically collected < 72 hours after admission to hospital, usuallycontainCA-MRSAclonesthatharborSCCmectype IVandproduceSSTI(Table1).Theseclonestendtobesen- sitivetomostantimicrobials,withtheexceptionoferythro- mycinandciprofloxacin,andcommonlyproducetheviru-lencefactorPVL.
recoMBination Between coMMunity anD hospital Mrsa clones
Thereisgrowingevidencethatvirulencegenescanbetrans-ferred between HA- and CA-MRSA strains. For example,
SCCmectypeIVtraits,usuallyassociatedwithCA-MRSA, havebeenobservedinnosocomialMRSAstrainswithsus-ceptibilitytofourormoreantimicrobials,67suggestingthat
CA-MRSA clones have been introduced into the hospital andarenowcirculatingasnosocomialpathogens.Similarly, biofilm-andenterotoxin-producing,non-multidrugresist-antMRSAthatdisplayedPFGEpatternssimilartoUSA800 causedseverenosocomialinfectionsintwoBrazilianhospi-talsin2007.68Clonesoriginatinginhospitalshavealsobeen
foundtobespreadinginthecommunity.InBrazil,MRSA strains isolated from the nasopharyngeal passages of chil-drenrecentlyadmittedtohospitalbetween2000and2001, werefoundtobemultidrugresistantandharboringSCCmec
typeIII.69
influence of Mrsa clones on Disease anD clinical practice
MRSA clones and nasal carriers
In a recent study, over 7% of hospital admissions were estimatedtobenasalcarriersofMRSA,70withthenares
potentially serving as a colonizing point and reservoir forfurtherinfection.WhilemostcarriersofS. aureusare thought to be colonized by a singleS. aureus clone, ap-proximately7%ofcarriersinonestudywerefoundtobe ‘discordant’carriers,colonizedbymorethanonestrain, andinsomecasescarrierswerecolonizedbyMRSAand MSSAstrainssimultaneously.71Indiscordantcarriers,the
presenceofmultiplestrainsofS. aureus presentstheop-portunity for horizontal genetic information exchange betweenstrains.71
MRSA clones isolated from the anterior nares can be varied, but are often community clones.70,72 In a homeless
population study in the USA,72 6.2% of individuals
car-riednasalMRSAclones,mostofwhichweretheCA-MRSA clonesUSA300andUSA1000.InchildrenwithinBrazilian day-carecenters,severalMRSAclonaltypes,includingSC-CmectypesIIIA,IVandV,havebeenidentified.73
Studies suggest that nasal carriers ofS. aureus may develop infection at a further site with the same clone. Forexample,inonestudy,>82%patientswhodeveloped bacteremia carried identicalS. aureus clones in the an-terior nares.74 Furthermore, clones present in the
ante-riornareswereshowntoappearinbacteremiainfections in the same patients up to 14 months later. In another study,>67%patientswithclinicalMRSAinfectionswere foundtocarryMRSAinthenares,andnasalcarriersof MRSA carried an enhanced risk of MRSA infections at othersitescomparedwiththeremainderofthepopula-tion.75 Healthcare workers are also at risk of becoming
carriers with the same clones that are present in hospi-talizedpatients,76andrisktransmittingthispathogento
MRSA clones and skin and soft tissue infections
MRSAisaprevalentandwidespreadcauseofSSTIs.InaUS study,MRSAwasthemostcommonpathogen-causingSSTI identified in emergency departments, with a prevalence of59%overall.48OftheMRSAisolates,72%wereasingle
strain,USA300-0114,whichharboredSCCmectypeIVand was positive for PVL. These isolates were associated with superficialinfectionsandabscesses,andwithdeeperinfec-tionsandosteomyelitis.48MRSAwasalsothemostcommon
pathogenassociatedwithSSTIsrequiringsurgicaldebride-mentina7-yearstudyinHouston.78
While most of the MRSA strains causing SSTIs are of community origin worldwide, differences in clonality and resistance pattern of MRSA strains between close geo- graphicareashavebeenreported.InCalifornia,forexam-ple,CA-MRSAaccountedfor93%ofSSTIsinSanFrancisco GeneralHospitalbetween2000and2002,butonlyfor69% inStanfordUniversityHospital;by2002,PVL-positiveST8-SCCmectypeIV(USA300)wasthepredominantstrainin San Francisco, showing sensitivity to most antimicrobials, whereasthemultidrugresistantcloneST5-SCCmectypeII (USA100)wasthepredominantclonecausingSSTIatStan-fordUniversityHospital.79
Thisstudyhighlightstheimpor-tance of local surveillance knowledge, even at the level of individualhospitalswithinthesamevicinity,whenchoosing antimicrobialtherapyforcommunity-basedSSTIs.
InLatinAmerica,limiteddataareavailabletodrawcon-clusions on the associations of clones with SSTIs. In the firstreportofcommunity-acquiredinfectionsintheregion, Ribeiroandcolleagues34observedPVL-positiveSCCmectype
IVCA-MRSAstrainsintwopatientswithSSTIandonewith septic arthritis, which corresponded to the Oceania South- westPacificclone(OSPC)fromAustralia,andsharedgeno-typiccharacteristicswithCA-MRSAstrainsintheUSAandin Europe.Asurveillancestudyconductedin2005inCordoba, Argentina,foundthat90%ofinfectionscausedbyCA-MRSA wereSSTIs,and89%werecausedbyCC5:ST5SCCmec type IVa-cstrains,35 whereasinCostaRica,clonesCC5:ST5SCC-mectypeIV(USA800)andCC8:SLV8SCCmectypeIV(USA 300) were found in community isolates from patients with SSTI,bothofwhichwerenegativeforPVL.29
MRSA clones and bacteremia
Data from the SENTRY Antimicrobial Surveillance Pro-graminArgentina,Brazil,Chile,Colombia,Venezuelaand Uruguay, have shown that MRSA is a common cause of nosocomialbacteremia,withaprevalenceof21.6%among bloodstreaminfectionisolates.80
MRSAnosocomialbacter- emiacausesseverecomplications,includinginfectiousen-docarditis,81andisassociatedwithelevatedmorbidityand
mortality.82 The selection of antibiotic treatment and the
durationoftreatmentforMRSAnosocomialbacteremiais oneofthemostcontroversialissuesinmedicine.81,82
BacteremiacanbecausedbybothCA-andHA-MRSA clones, and the severity of disease is dependent on clonal type.Inarecentstudy,83SCCmectypeIIHA-MRSAclones
werefoundtocausehighermortalitythanSCCmectypeIVa CA-MRSAclones,whereasSCCmectypeIVaclonescaused greater metastatic infection. Also, while the majority of community-acquired and hospital-acquired bacteremias werecausedbySCCmectypeIVaandSCCmectypeIIMRSA clones,respectively,SCCmectypeIVawaspresentinsome hospital-acquired bacteremias, and SCCmec type II was presentinsomecommunity-acquiredbacteremias.83
MRSA clones and infectious endocarditis
MRSA is a common pathogen in infectious endocarditis (IE),withprevalenceofoveronethirdinsomecountries, includingBrazil(37.5%)andtheUSA(37.2%).81S. aureus
IEisusuallyacquiredoutsidehospital,butpredominatesas ahealthcare-associatedinfection,whichaccountsfor54%of casesinBrazil.81ThefirstreportedcaseofIEinBrazilcaused
byCA-MRSAwaspublishedin2008,84andwasattributedto
MRSAharboringSCCmectypeIVandpositiveforPVL.In Korea,aPVL-negativeCA-MRSAcloneST72SCCmecIVa hasbeenidentifiedasthecauseofIEinfectionsinpreviously healthyindividualswithnoreportedriskfactorsforIE.85
conclusion: current challenges anD future Directions
Molecular epidemiological studies have highlighted the continuing global evolution and spread of MRSA clones with increased resistance to antimicrobial drugs and en- hancedvirulence.Thefactorscontributingtothedissemi-nationofMRSAclonesareonlypartiallyunderstood,but are thought to include the migration of human popula-tions, ineffective methods to control transmission of MRSAfrominfectedpatients,andtreatmentstrategies,in-cludingtheinappropriateuseandchoiceofantibiotics.86,87
Inhospitals,patientsalreadycarryingMRSAwhenadmit-ted are at greater risk of developing an infection derived fromthecolonizedbacteria,oroftransmittingMRSAto otherpatients.
InLatinAmerica,pandemicclonesarecommonplacein hospitalsacrosstheregion,andcommunity-associatedin-fectionsaregrowinginnumber.1
Clonescirculatinginthe regionshowgeneticdiversity,althoughcommongenesen-codingmultidrugresistancetoantimicrobialsareexpressed. Enhanced pathogenic properties, including production ofbiofilmsandproductionofenterotoxins,havebeende-scribedforcertainclones,andtheabilityofnosocomialand communityclonestointerchangegeneticmaterialhasalso beenidentified.
However, surveillance data for specific MRSA clones within the region is limited,1
centers.Paradoxically,countriesshowingthehighestpreva-lence of nosocomial MRSA infections, such as Peru, often provideverylittleinformationwithregardtomolecularepi- demiologyandclinicaloutcome.Regionalsurveillancepro-grams,usingcentralreferencelaboratoriesandintegrating informationfromhealthcenterswithdifferentcomplexities, arerequiredifwearetounderstandmorefullythedevelop-ingpatternofMRSAinfectionsacrossLatinAmerica,andto designbettertreatmentandpreventionstrategies.
iMplications for clinical practice
•ThecharacteristicsofdifferentMRSAclonesare associatedwithclinicalpresentationandoutcomes.
• ClonaldisseminationisrapidforbothCA-MRSAandHA-MRSA,andrecombination betweenCAandHAclonesistakingplace.
•Regionalmolecularepidemiologyprogramsare requiredtounderstandmorefullythedeveloping patternofMRSAinfectionsacrossLatinAmerica.
•Moreaccurateinformationontheprecise distributionofclonesintheregionmay helpcliniciansinunderstandingtheriskof infectionbycertainclonesandinchoosing appropriateempiricantimicrobialtherapy.
acknowleDgeMents
Financial support
PfizerInc.,NewYork,NY,USA,providedsupportformeet- ingsoftheLatinAmericanWorkingGrouponGramPosi- tiveResistance.PfizerInc.hadnoinvolvementinthecollec-tion, analysis and interpretation of data, in the writing of themanuscripts,orinthedecisiontosubmitthearticlesfor publication.
Manuscript preparation
The support provided by Choice Pharma (Hitchin, UK), funded by Pfizer Inc., consisted of manuscript formatting andwritingassistance.
Disclosures
E. Rodríguez-Noriega:Advisory Board member for Pfizer; consultant for Pfizer,Wyeth, Johnson & Johnson and No-vartis;receivedresearchgrantsfromPfizer,Wyeth,Johnson &Johnson,Schering-PloughandCerexa.
C. Seas: Advisory Board member and consultant for Pfizer;receivedresearchfundingfromTherevance,Cerexa, Schering-PloughandAvexa.
references
1. Guzmán-BlancoM,MejíaC,IsturizRet al.Epidemiologyof
methicillin-resistantStaphylococcus aureus (MRSA) in Latin
America.IntJAntimicrobAgents.2009;34(4):304-8.
2. Rodríguez-Noriega E, Seas C, Guzmán-Blanco Met al.
Evo-lutionofmethicillin-resistantStaphylococcus aureusclonesin LatinAmerica.IntJInfectDis.2010;14:e560-6.
3. Ito T, Ma XX, Takeuchi Fet al. Novel typeV staphylococcal cassette chromosome mec driven by a novel cassette chro-mosome recombinase, ccrC. Antimicrob Agents Chemother. 2004;48(7):2637-51.
4. JevonsMP,CoeAW,ParkerMT.Methicillinresistanceinsta-phylococci.Lancet.1963;1(7287):904-7.
5. Crisostomo MI, Westh H, Tomasz Aet al. The evolution of
methicillin resistance inStaphylococcus aureus: similarity of
geneticbackgroundsinhistoricallyearlymethicillin-suscepti-bleand-resistantisolatesandcontemporaryepidemicclones. ProcNatlAcadSciUSA.2001;98(17):9865-70.
6. DeurenbergRH,StobberinghEE.Themolecularevolutionof hospital-andcommunity-associatedmethicillin-resistantSta-phylococcus aureus.CurrMolMed.2009;9(2):100-15. 7.
DeurenbergRH,StobberinghEE.TheevolutionofStaphyloco-ccus aureus.InfectGenetEvol.2008;8(6):747-63.
8. DonnioPY,FevrierF,BifaniPet al.Molecularandepidemio-logical evidence for spread of multiresistant methicillin-sus-ceptibleStaphylococcus aureusstrainsinhospitals.Antimicrob AgentsChemother.2007;51(12):4342-50.
9. CooksonBD,RobinsonDA,MonkABet al.Evaluationofmo-lecular typing methods in characterizing a European
collec-tion of epidemic methicillin-resistantStaphylococcus aureus
strains: the HARMONY collection. J Clin Microbiol. 2007; 45(6):1830-7.
10. Naimi TS, LeDell KH, Como-Sabetti Ket al. Comparison of
community- and health care-associated methicillin-resistant Staphylococcus aureusinfection.JAMA2003;290(22):2976-84. 11. GorwitzRJ.Understandingthesuccessofmethicillin-resistant
Staphylococcus aureusstrainscausingepidemicdiseaseinthe community.JInfectDis.2008;197(2):179-82.
12. FlynnN,CohenSH.ThecontinuingsagaofMRSA.JInfect Dis.2008;197(9):1217-9.
13.
AyliffeGA.Theprogressiveintercontinentalspreadofmethi-cillin-resistantStaphylococcus aureus. Clin Infect Dis. 1997;
24(Suppl1):S74-9.
14. Robinson DA, Enright MC. Multilocus sequence typing and
the evolution of methicillin-resistantStaphylococcus aureus.
ClinMicrobiolInfect.2004;10(2):92-7.
15. DenisO,DeplanoA,NonhoffCet al.Nationalsurveillanceof methicillin-resistantStaphylococcus aureusinBelgianhospitals indicatesrapiddiversificationofepidemicclones.Antimicrob AgentsChemother.2004;48(9):3625-9.
16. Aires-de-Sousa M, Correia B, de Lencastre H. Changing pat- ternsinfrequencyofrecoveryoffivemethicillin-resistantSta-phylococcus aureusclonesinPortuguesehospitals:surveillance overa16-yearperiod.JClinMicrobiol.2008;46(9):2912-7. 17. OliveiraDC,TomaszA,deLencastreH.Secretsofsuccessof
ahumanpathogen:molecularevolutionofpandemicclones ofmeticillin-resistantStaphylococcus aureus.LancetInfectDis. 2002;2(3):180-9.
19. PotelC,AlvarezM,AlvarezPet al.Evolution,antimicrobialsus- ceptibilityandassignmenttointernationalclonesofmethicillin-resistantStaphylococcus aureus isolated over a 9-year period in twoSpanishhospitals.ClinMicrobiolInfect.2007;13(7):728-30. 20. PopovichKJ,WeinsteinRA,HotaB.Arecommunity-associat-edmethicillin-resistantStaphylococcus aureus(MRSA)strains replacing traditional nosocomial MRSA strains? Clin Infect Dis.2008;46(6):787-94.
21. SaderHS,PignatariAC,HollisRJ,JonesRN.Evaluationofin-terhospital spread of methicillin-resistantStaphylococcus au-reusinSaoPaulo,Brazil,usingpulsed-fieldgelelectrophoresis ofchromosomalDNA.InfectControlHospEpidemiol.1994; 15(5):320-3.
22. AiresdeSousaM,MiragaiaM,SanchesISet al.Three-yearas-sessmentofmethicillin-resistantStaphylococcus aureusclones inLatinAmericafrom1996to1998.JClinMicrobiol.2001; 39(6):2197-205.
23. SolaC,GribaudoG,VindelAet al.Identificationofanovel methicillin-resistantStaphylococcus aureus epidemic clone in Cordoba,Argentina,involvedinnosocomialinfections.JClin Microbiol.2002;40(4):1427-35.
24. Velazquez-MezaME,AiresdeSousaM,Echaniz-AvilesGet al. Surveillanceofmethicillin-resistantStaphylococcus aureusina pediatrichospitalinMexicoCityduringa7-yearperiod(1997 to2003):clonalevolutionandimpactofinfectioncontrol.J ClinMicrobiol.2004;42(8):3877-80.
25. Akpaka PE, Kissoon S, Rutherford Cet al. Molecular epide-miologyofmethicillin-resistantStaphylococcus aureusisolates from regional hospitals in Trinidad and Tobago. Int J Infect Dis.2007;11(6):544-8.
26. Amaral MM, Coelho LR, Flores RPet al. The predominant variantoftheBrazilianepidemicclonalcomplexofmethicil-lin-resistantStaphylococcus aureushasanenhancedabilityto producebiofilmandtoadheretoandinvadeairwayepithelial cells.JInfectDis.2005;192(5):801-10.
27. CruzC,MorenoJ,RenzoniAet al.Trackingmethicillin-resist-antStaphylococcus aureusclonesinColombianhospitalsover 7years(1996-2003):emergenceofanewdominantclone.Int JAntimicrobAgents.2005;26(6):457-62.
28. Echaniz-Aviles G,Velazquez-Meza ME, Aires-de-Sousa Met al. Molecular characterisation of a dominant methicillin-re-sistantStaphylococcus aureus(MRSA)cloneinaMexicanhos-pital(1999-2003).ClinMicrobiolInfect.2006;12(1):22-8. 29. GoeringRV,ShawarRM,ScangarellaNEet
al.Molecularepi-demiologyofmethicillin-resistantandmethicillin-susceptible Staphylococcus aureusisolatesfromglobalclinicaltrials.JClin Microbiol.2008;46(9):2842-7.
30. GomesAR,SanchesIS,AiresdeSousaMet
al.Molecularepi-demiology of methicillin-resistantStaphylococcus aureus in
Colombian hospitals: dominance of a single unique multid-rug-resistantclone.MicrobDrugResist.2001;7(1):23-32. 31. RamirezBarbaEJ,RosenthalVD,HigueraFet
al.Device-asso-ciatednosocomialinfectionratesinintensivecareunitsinfour Mexicanpublichospitals.AmJInfectControl.2006;34(4):244-7.
32. Mayor L, Ortellado J, Menacho Cet al. Molecular
charac-terization of methicillin-resistantStaphylococcus aureus
iso-latescollectedinAsuncion,Paraguay.JClinMicrobiol.2007; 45(7):2298-300.
33. Melo MC, Silva-Carvalho MC, Ferreira RLet al. Detection
and molecular characterization of a gentamicin-susceptible,
methicillin-resistantStaphylococcus aureus (MRSA) clone in
RiodeJaneirothatresemblestheNewYork/Japaneseclone.J HospInfect.2004;58(4):276-85.
34. Ribeiro A, Dias C, Silva-Carvalho MCet al. First report of
infectionwithcommunity-acquiredmethicillin-resistantSta-phylococcus aureusinSouthAmerica.JClinMicrobiol.2005; 43(4):1985-8.
35. Sola C, Saka HA, Vindel A, Bocco JL. Emergence and dis-semination of a community-associated methicillin-resistant
Panton-Valentine leucocidin-positiveStaphylococcus aureus
clonesharingthesequencetype5lineagewiththemostpreva-lentnosocomialcloneinthesameregionofArgentina.JClin Microbiol.2008;46(5):1826-31.
36. ReyesJ,RinconS,DiazLet al.Disseminationofmethicillin-resistantStaphylococcus aureusUSA300sequencetype8line-ageinLatinAmerica.ClinInfectDis.2009;49(12):1861-7.
37. Ma XX, Galiana A, Pedreira Wet al. Community-acquired
methicillin-resistantStaphylococcus aureus, Uruguay. Emerg
InfectDis.2005;11(6):973-6.
38. Ramos RL, Teixeira LA, Ormonde LRet al. Emergence of
mupirocin resistance in multiresistantStaphylococcus aureus
clinicalisolatesbelongingtoBrazilianepidemiccloneIII::B:A. JMedMicrobiol.1999;48(3):303-7.
39. CorsoA,SantosSanchesI,AiresdeSousaMet al.Spreadof a methicillin-resistant and multiresistant epidemic clone of Staphylococcus aureusinArgentina.MicrobDrugResist.1998; 4(4):277-88.
40. SeasC,HernandezK,RamosRet al.Oxacillin-resistantand multidrug-resistantStaphylococcus aureusinLima,Peru.Infect ControlHospEpidemiol.2006;27(2):198-200.
41. PfizerVenezuela S.A.Venezuelan program of surveillance of bacterial resistance to antibiotics; 1998-2006. (http://www. provenra.org)Accessed19May2010.
42. AylwinM,SerriM,GracíaPet al.Staphylococcus aureusmeti-cilinoresistenteasociadoalacomunidadenChile(A-C021). AbstractpresentedattheXXIVChileanConferenceofInfec-tiousDiseases.Pucón,Chile,2007.
43. Noríega LM, Gonzalez P, Hormazabal JCet al. Community
acquired infections with methicillin resistant strains of
Sta-phylococcus aureus: report of five cases. Rev Med Chil. 2008; 136(7):885-91.
44. DiepBA,CarletonHA,ChangRFet al.Rolesof34virulence genesintheevolutionofhospital-andcommunity-associated strainsofmethicillin-resistantStaphylococcus aureus.JInfect Dis.2006;193(11):1495-503.
45. DiepBA,Palazzolo-BallanceAM,TattevinPet al.Contribu-tionofPanton-Valentineleukocidinincommunity-associated methicillin-resistantStaphylococcus aureuspathogenesis.PLoS One.2008;3(9):e3198.
46. Diep BA, Sensabaugh GF, Somboona NSet al. Widespread
skin and soft-tissue infections due to two
methicillin-re-sistantStaphylococcus aureus strains harboring the genes
for Panton-Valentine leucocidin. J Clin Microbiol. 2004; 42(5):2080-4.
47. FrancisJS,DohertyMC,LopatinUet al.Severecommunity-
onsetpneumoniainhealthyadultscausedbymethicillin-re-sistantStaphylococcus aureus carrying the Panton-Valentine
leukocidingenes.ClinInfectDis.2005;40(1):100-7.
48. MoranGJ,KrishnadasanA,GorwitzRJet al.Methicillin-re-sistantS. aureusinfectionsamongpatientsintheemergency department.NEnglJMed.2006;355(7):666-74.
49. LimbagoB,FosheimGE,SchoonoverVet
al.Characteriza-tion of methicillin-resistantStaphylococcus aureus isolates
50. McCaskill ML, Mason EO, Jr., Kaplan SLet al. Increase of the USA300 clone among community-acquired methicillin-susceptibleStaphylococcus aureus causing invasive infections. PediatrInfectDisJ.2007;26(12):1122-7.
51. LinaG,PiemontY,Godail-GamotFet al.InvolvementofPan-ton-Valentineleukocidin-producingStaphylococcus aureusin primaryskininfectionsandpneumonia.ClinInfectDis.1999; 29(5):1128-32.
52. GilletY,IssartelB,VanhemsPet al.AssociationbetweenSta-phylococcus aureusstrainscarryinggeneforPanton-Valentine leukocidinandhighlylethalnecrotisingpneumoniainyoung immunocompetentpatients.Lancet.2002;359(9308):753-9. 53. TristanA,BesM,MeugnierHet
al.GlobaldistributionofPan-ton-Valentine leukocidin-positive methicillin-resistantStaphy-lococcus aureus,2006.EmergInfectDis.2007;13(4):594-600. 54. VivoniAM,DiepBA,deGouveiaMagalhaesACet al.Clonal
composition ofStaphylococcus aureus isolates at a Brazilian
universityhospital:identificationofinternationalcirculating lineages.JClinMicrobiol.2006;44(5):1686-91.
55. SolaC,SakaHA,VindelA,BoccoJL.HighfrequencyofPan- ton-Valentineleukocidingenesininvasivemethicillin-suscep-tibleStaphylococcus aureus strains and the relationship with methicillin-resistantStaphylococcus aureusinCordoba,Argen-tina.EurJClinMicrobiolInfectDis.2007;26(4):281-6.
56. Souza RR, Coelho LR, Botelho AMet al. Biofilm formation
andprevalenceoflukF-pv,seb,secandtstgenesamonghos-pital-andcommunity-acquiredisolatesofsomeinternational
methicillin-resistant Staphylococcus aureus lineages. Clin
MicrobiolInfect.2009;15(2):203-7.
57. Rice LB. Antimicrobial resistance in gram-positive bacteria. AmJMed.2006;119(6Suppl1):S11-9;discussionS62-70. 58. DeurenbergRH,VinkC,KalenicSet
al.Themolecularevo-lution of methicillin-resistantStaphylococcus aureus. Clin
MicrobiolInfect.2007;13(3):222-35.
59. ChangS,SievertDM,HagemanJCet al.Infectionwithvan-comycin-resistantStaphylococcus aureuscontainingthevanA resistancegene.NEnglJMed.2003;348(14):1342-7.
60. TenoverFC,MoelleringRC,Jr.Therationaleforrevisingthe ClinicalandLaboratoryStandardsInstitutevancomycinmini- malinhibitoryconcentrationinterpretivecriteriaforStaphylo-coccus aureus.ClinInfectDis.2007;44(9):1208-15.
61. Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH.
Practicaldiskdiffusionmethodfordetectionofinducibleclin-damycin resistance inStaphylococcus aureus and
coagulase-negativestaphylococci.JClinMicrobiol.2003;41(10):4740-4.
62. Patel M,Waites KB, Moser SAet al. Prevalence of inducible
clindamycinresistanceamongcommunity-andhospital-asso-ciatedStaphylococcus aureus isolates. J Clin Microbiol. 2006; 44(7):2481-4.
63. Chavez-Bueno S, Bozdogan B, Katz Ket al. Inducible
clin-damycin resistance and molecular epidemiologic trends of pediatriccommunity-acquiredmethicillin-resistantStaphylo-coccus aureusinDallas,Texas.AntimicrobAgentsChemother. 2005;49(6):2283-8.
64. VivoniAM, Santos KR, de-Oliveira MPet al. Mupirocin for controllingmethicillin-resistantStaphylococcus aureus:lessons fromadecadeofuseatauniversityhospital.InfectControl HospEpidemiol.2005;26(7):662-7.
65. SafdarN,BradleyEA.Theriskofinfectionafternasalcoloniza-tionwithStaphylococcus aureus.AmJMed.2008;121(4):310-5. 66. SaimanL,CronquistA,WuFet al.Anoutbreakofmethicil-lin-resistantStaphylococcus aureusinaneonatalintensivecare unit.InfectControlHospEpidemiol.2003;24(5):317-21.
67. deATrindadeP,PachecoRL,CostaSFet al.PrevalenceofSC-
CmectypeIVinnosocomialbloodstreamisolatesofmethi-cillin-resistantStaphylococcus aureus. J Clin Microbiol. 2005;
43(7):3435-7.
68. de Miranda OP, Silva-Carvalho MC, Ribeiro Aet al.
Emer-gence in Brazil of methicillin-resistantStaphylococcus aureus isolatescarryingSCCmecIVthatarerelatedgeneticallytothe USA800clone.ClinMicrobiolInfect.2007;13(12):1165-72.
69. Lamaro-Cardoso J, Castanheira M, de Oliveira RMet al.
Carriage of methicillin-resistant Staphylococcus aureus
in children in Brazil. Diagn Microbiol Infect Dis. 2007; 57(4):467-70.
70. HidronAI,KourbatovaEV,HalvosaJSet al.Riskfactorsfor
colonization with methicillin-resistantStaphylococcus aureus
(MRSA)inpatientsadmittedtoanurbanhospital:emergence of community-associated MRSA nasal carriage. Clin Infect Dis.2005;41(2):159-66.
71. Cespedes C, Said-Salim B, Miller Met al. The
clonal-ityofStaphylococcus aureusnasalcarriage.JInfectDis.2005; 191(3):444-52.
72. Pan ES, Diep BA, Charlebois EDet al. Population dynamics
ofnasalstrainsofmethicillin-resistantStaphylococcus aureus–
andtheirrelationtocommunity-associateddiseaseactivity.J InfectDis.2005;192(5):811-8.
73. Lamaro-CardosoJ,deLencastreH,KipnisAet al.Molecular
epidemiologyandriskfactorsfornasalcarriageofStaphylo-coccus aureus and methicillin-resistantS. aureus in infants
attending day-care centers in Brazil. J Clin Microbiol. 2009; 47(12):3991-7.
74. vonEiffC,BeckerK,MachkaKet al.Nasalcarriageasasource
of Staphylococcus aureus bacteremia. Study Group. N Engl J
Med.2001;344(1):11-6.
75. RobicsekA,SusenoM,BeaumontJLet al.Predictionofme-thicillin-resistantStaphylococcus aureusinvolvementindisease sitesbyconcomitantnasalsampling.JClinMicrobiol.2008; 46(2):588-92.
76. CespedesC,MillerM,QuagliarelloBet al.Differencesbetween Staphylococcus aureus isolates from medical and nonmedical hospitalpersonnel.JClinMicrobiol.2002;40(7):2594-7. 77. EveillardM,MartinY,HidriNet
al.Carriageofmethicillin-resistantStaphylococcus aureus among hospital employees:
prevalence,duration,andtransmissiontohouseholds.Infect ControlHospEpidemiol.2004;25(2):114-20.
78. AwadSS,ElhabashSI,LeeLet
al.Increasingincidenceofme-thicillin-resistantStaphylococcus aureus skin and soft-tissue
infections: reconsideration of empiric antimicrobial therapy. AmJSurg.2007;194(5):606-10.
79. Bhattacharya D, Carleton H, Tsai CJet al. Differences in
clinical and molecular characteristics of skin and soft tissue
methicillin-resistantStaphylococcus aureus isolates between
twohospitalsinNorthernCalifornia.JClinMicrobiol.2007; 45(6):1798-803.
80. BiedenbachDJ,MoetGJ,JonesRN.Occurrenceandantimi-crobial resistance pattern comparisons among bloodstream infection isolates from the SENTRY Antimicrobial Surveil-lanceProgram(1997-2002).DiagnMicrobiolInfectDis.2004; 50(1):59-69.
81. FowlerVG,Jr.,MiroJM,HoenBet al.Staphylococcus aureus endocarditis:aconsequenceofmedicalprogress.JAMA.2005; 293(24):3012-21.
82. Corey GR.Staphylococcus aureus bloodstream infections:
83. GangaR,RiedererK,SharmaMet al.RoleofSCCmectypein outcomeofStaphylococcus aureusbacteremiainasinglemedi-calcenter.JClinMicrobiol.2009;47(3):590-5.
84. FortesCQ,EspanhaCA,BustorffFPet al.Firstreportedcaseof infectiveendocarditiscausedbycommunity-acquiredmethicil-lin-resistantStaphylococcus aureus not associated with health-carecontactinBrazil.BrazJInfectDis.2008;12(6):541-3. 85. LeeSY,KimJY,KimJHet
al.Acaseofprimaryinfectiveendo-carditiscausedbycommunity-associatedmethicillin-resistant Staphylococcus aureusinahealthyindividualandcolonization inthefamily.YonseiMedJ.2009;50(1):152-5.
86. AiresdeSousaM,SanchesIS,FerroMLet al.Intercontinental spreadofamultidrug-resistantmethicillin-resistantStaphylo-coccus aureusclone.JClinMicrobiol.1998;36(9):2590-6. 87. ShopsinB,MathemaB,ZhaoXet al.Resistanceratherthan