revbrashematolhemoter.2015;37(2):142–143
w w w . r b h h . o r g
Revista
Brasileira
de
Hematologia
e
Hemoterapia
Brazilian
Journal
of
Hematology
and
Hemotherapy
Letter
to
the
Editor
Is
the
BCR-ABL/GUSB
transcript
level
at
diagnosis
an
early
predictive
marker
for
chronic
myeloid
leukemia
patients
treated
with
imatinib?
DearEditor,
Thedevelopmentofthefirst target-specifictyrosinekinase inhibitor(TKI)anditsintroductionintheclinicalpractice rad-ically changed chronic myeloid leukemia (CML) treatment. MonitoringtherapeuticresponsetoTKIsisacriticalstepin themanagementofCML.1
Recently,severalfollow-upstudiesuponwhichthe Euro-peanLeukemiaNet2013(ELN)recommendationswerebased, pointedtotheimportanceofearlyclearanceofleukemiccells as demonstrated by molecular methods. Attaining a BCR-ABLIStranscriptlevel≤10%threemonthsafterinitialimatinib mesylate (IM) treatment was found to be associated with afavorableoutcome,includinglongerprogression-free(PFS) andoverallsurvival(OS),andhigherprobabilityofachieving completecytogeneticresponse(CCyR)and majormolecular response(MMR).1
Quantification of the BCR-ABL transcript level reflects leukemicburden,andiscarriedoutbyquantitativereal-time polymerase chainreaction(RQ-PCR). Molecular response is basedon theratio ofBCR-ABL transcriptlevelsand a con-trolgene.Resultsareexpressedaccordingtoaninternational scale(IS)assignedtoeverypatientatdiagnosis,whichisequal to 100%of BCR-ABL/controlgene transcripts, regardless of theabsoluteamountofBCR-ABLtranscripts.Thus,theactual leukemicburden ofpatientsatdiagnosis is nottaken into account.2
Anidealcontrolgenewouldbeexpectedtobeuniformly expressedindifferentcelltypesregardlessofitsproliferative statusaswellasbeunaffectedbytherapeuticregimens, con-stantbetweenindividualsandexpressedatalevelsimilarto BCR-ABL.Infact,this controlgenedoesnotexist, andBCR andABLarethemostwidelyusedcontrolgenesfor quanti-fyingBCR-ABLtranscripts,mainlyduetohistoricalreasons. However,bothBCRandABLcontrolgenesdonotshow linear-itywithBCR-ABLtranscriptlevelsabove10%contrarytothe GUSBgenethatisnotaffectedbyhigh-leveldistortionswhich allowforbetterestimationsoftheBCR-ABLtranscriptlevelat diagnosis.3
In this study, the BCR-ABL transcript levels of 31 CML patients under IM treatment were analyzed by RQ-PCR in
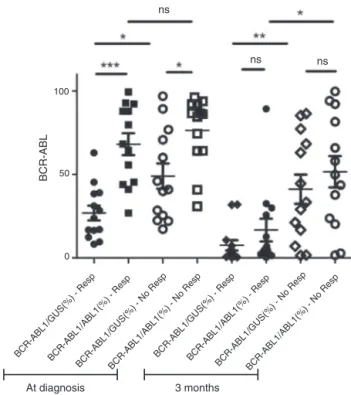
respecttotheABLandGUSBcontrolgenesatdiagnosisand afterthreemonthsoftherapy.Thesepatientswerefollowed up foratleast 24 months.Themedian BCR-ABL1/ABL and BCR-ABL1/GUSB transcript levels at diagnosis were 87.84% (range:16.24–184.4)and29.8%(range:5.76–216.9),respectively. Atthree months,the medianBCR-ABL/ABLtranscriptlevel was7.14(range:0.053–307)whereasthemedianBCR-ABL/GUS transcript levelwas 21.94(range: 0.19–85.16). Patients were classified asoptimal responders ornon-responders (failure ofresponse)accordingtoaBCR-ABL1IStranscriptlevel≤0.1%
and >0.1% at 12 months. In responders, the median BCR-ABL/ABLandBCR-ABL/GUStranscriptlevelsatdiagnosiswere 68.13%(range:26.8–99.29)and23.77(range:8.2–62.97), respec-tivelywhileinnon-responderstheselevelswere86.10(range: 30.87–96.11)and40.92(range:17.21–96.85).
The median BCR-ABL/ABL of responders and non-responders at diagnosis was not significantly different (p-value=0.89) while the median BCR-ABL/GUS between responders and non-responders at diagnosis was signifi-cantlydifferent(p-value<0.001)indicatingthat,unlikeABL, GUS levels are capable of discriminating responders from non-responders(Figure1).Themediantranscriptlevelof BCR-ABL/GUSofrespondersatdiagnosiswas28.38%whichmight beconsideredathresholdforearlydiscriminationaspatients withlevelsunder28.38%werelesslikelytoachieveMMRat12 months(p-value<0.05).
Comparisonsoftranscriptlevelsthreemonthsafter ini-tiating IM treatment were also carried out considering a BCR-ABL/controlgeneIS thresholdof≤10%asdiscriminative
ofrespondersvs.non-responders.Inpatientsconsideredtobe optimalresponders,themedianofthetranscriptsestimated withanycontrolgenewasbelow10%,asexpectedaccordingto previousreports.Wedidnotobserveastatisticallysignificant differencebetweenABLandGUScontrolgenes(p-value=0.19) ineitherrespondersor non-responders(p=0.41),indicating thatatthethreemonthtimepoint,bothgenescanbeequally usedaspredictivebiomarkers,showingsignificantdifferences betweenrespondersandnon-responders(p-value=0.003for GUSandp-value=0.01forABL;Figure1).
revbrashematolhemoter.2015;37(2):142–143
143
ns
ns
100
50
BCR-ABL
0
At diagnosis 3 months
BCR-AB
L1/GUS(%) - Resp
BCR-A BL1/ABL1(%) - R
esp
BCR -ABL1/GUS(%) -
Res p
BCR-ABL1/ ABL1(%) -
Resp
BCR-ABL1
/GUS(%) - No Resp
BCR-ABL1/AB L1(%) -
No Resp
BCR-ABL1/G US(%) -
No Resp
BCR-ABL1/ABL1 (%) -
No Res p
ns
Figure1–Themedianlevelsofthetranscriptsconsidering bothcontrolgenes(GUSandABL)atdiagnosisandafter threemonthsonimatinib(IM)asfirstlinetherapy.Patients arediscriminatedasrespondersornon-responders accordingtotheirresponseat12monthsaccordingtothe ELNguidelines.Themediansofthetranscriptswere comparedbyMann–Whitneyunpairedtwo-tailedtest.At diagnosis:*p-value<0.001.At3months:*p-value=0.01; **p-value=0.003.Resp=responders;No
Resp=non-responders;ns=non-significant.
futureresponse. Inthis study,we usedparameters recom-mendedforidentifyingoptimalresponse(withBCR-ABLIS≤10
atthreemonthsand≤0.1at12monthsafterinitiating treat-ment).However,BCR-ABL/ABLtranscriptlevelscannotbeused aspredictiveestimatesduetothelackoflinearityoftheABL geneinassessingleukemicburdenatlevelsabove10%and actuallevelsofBCR-ABLtranscriptsatdiagnosiscanbe accu-ratelyestimatedwithGUSasthecontrolgene.Wefoundthat highlevelsofBCR-ABL/GUSatdiagnosiswereassociatedwith a lower probability ofachieving optimalresponse (p-value <0.001)andlowratesofCyCRafter12monthsofIMtherapy (p-value<0.001).ThesefindingscoincidedwithVignerietal.4who
showedthathighratesofBCR-ABL/GUSwereassociatedwitha lowprobabilityofevent-freesurvival(p-value<0.001)andPFS
(p-value=0.01).Asexpected,thelossofABLlinearity result-ingintranscriptquantificationwithhighlevelsofleukemic burdenindicatedthatABL,ifusedasthecontrolgeneat diag-nosis,wouldnotprovidepredictiveestimates.Conversely,the useofGUSasthecontrolgeneallowsforareliableprediction oftherapeuticresponsebasedonBCR-ABLtranscriptlevelsat diagnosis.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest.
r
e
f
e
r
e
n
c
e
s
1.BaccaraniM,DeiningerMW,RostiG,HochhausA,SoveriniS,
ApperleyJF,etal.EuropeanLeukemiaNetrecommendations
forthemanagementofchronicmyeloidleukemia:2013.Blood.
2013;122(6):872–84.
2.GabertJ,BeillardE,vanderVeldenVH,BiW,GrimwadeD,
PallisgaardN,etal.Standardizationandqualitycontrolstudies
ofreal-timequantitativereversetranscriptasepolymerase
chainreactionoffusiongenetranscriptsforresidualdisease
detectioninleukemia–aEuropeAgainstCancerprogram.
Leukemia.2003;17(12):2318–57.
3.CrossNC.Standardisationofmolecularmonitoringforchronic
myeloidleukaemia.BestPractResClinHaematol.
2009;22(3):355–65.
4.VigneriPG,StagnoF,StellaS,CupriA,ForteS,MassiminoM,
etal.HighBCR-ABL/GUSEILevelsatdiagnosisareassociated
withunfavorableresponsestoimatinib.Blood.2013;122(21),
abstract#1495.
SimoneBoneckera,MarinaMagnagob,JaspalKaedac,
CristianaSolzab, IlanaZalcbergRenaulta,∗
aInstitutoNacionaldoCâncer(INCA),RiodeJaneiro,RJ,Brazil bUniversidadedoEstadodoRiodeJaneiro(UERJ),RiodeJaneiro,RJ,
Brazil
cUniversitätsmedizinBerlin,Berlin,Germany
∗Correspondingauthorat:Prac¸aCruzVermelha,23,6◦ andar,
LaboratóriosCEMO,Centro,20230-130RiodeJaneiro,RJ,Brazil. E-mailaddress:zalcberg@inca.gov.br(I.ZalcbergRenault).
Received14July2014 Accepted27August2014 Availableonline31January2015
http://dx.doi.org/10.1016/j.bjhh.2014.08.003