ContentslistsavailableatSciVerseScienceDirect
Journal
of
Clinical
Virology
j ourna l h o me p a g e :w w w . e l s e v i e r . c o m / l o c a t e / j c vGenomic
signatures
and
antiviral
drug
susceptibility
profile
of
A(H1N1)pdm09
Marta
Tiago
Gíria
a, Helena
Rebelo
de
Andrade
a,b,∗, Luís
André
Santos
b, Vanessa
Martins
Correia
b,
Sónia
Vicente
Pedro
c,
Madalena
Almeida
Santos
daCPM-URIA,FaculdadedeFarmáciadaUniversidadedeLisboa,Lisboa,Portugal
bDepartamentodeDoenc¸asInfecciosas,InstitutoNacionaldeSaúdeDrRicardoJorge,Lisboa,Portugal
cUnidadedeTecnologiaeInovac¸ão,DepartamentodeGenética,InstitutoNacionaldeSaúdeDrRicardoJorge,Lisboa,Portugal dServic¸odePatologiaClínica,Virologia/BiologiaMolecular,HospitaldeCurryCabral,Lisboa,Portugal
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received8July2011 Receivedinrevisedform 20September2011 Accepted8November2011 Keywords: A(H1N1)pdm09 Viralfitness Virulence Internalproteins Antiviraldrugsusceptibility Riskassessment
a
b
s
t
r
a
c
t
Background:Geneticchangesininfluenzasurfaceandinternalgenescanalterviralfitnessandvirulence. MutationtrendanalysisandantiviraldrugsusceptibilityprofilingofA(H1N1)pdm09virusesisessential forriskassessmentofemergentstrainsanddiseasemanagement.
Objective:ToprofilegenomicsignaturesandantiviraldrugresistanceofA(H1N1)pdm09virusesandto discussthepotentialroleofmutatedresiduesinhumanhostadaptationandvirulence.
Studydesign:A(H1N1)pdm09virusescirculatinginPortugalduringpandemicandpost-pandemicperiods and2009/2010season.ViruseswereisolatedinMDCK-SIAT1cellcultureandsubjectedtomutation analysisofsurfaceandinternalproteins,andtoantiviraldrugsusceptibilityprofiling.
Results:TheA(H1N1)pdm09strainscirculatingduringtheepidemicperiodinPortugalwereresistant toamantadine.Themajorityofthestrainswerefoundtobesusceptibletooseltamivirandzanamivir, withfiveoutlierstoneuraminidaseinhibitors(NAIs)identified.Specificmutationpatternsweredetected withinthefunctionaldomainsofinternalproteinsPB2,PB1,PA,NP,NS1,M1andNS2/NEP,whichwere commontoallisolatesandalsosomecluster-specific.
Discussion:Modificationofviralgenometranscription,replicationandapoptosiskinetics,changesin antigenicityandantiviraldrugsusceptibilityareknowndeterminantsofvirulence.Wereportseveral pointmutationswithputativerolesinviralfitnessandvirulence,anddiscusstheirpotentialtoresultin morevirulentphenotypes.Monitoringofspecificmutationsandgeneticpatternsininfluenzaviralgenes isessentialforriskassessingemergentstrains,diseaseepidemiologyandpublichealthimplications.
© 2011 Elsevier B.V. All rights reserved.
1. Background
Theemergence of virulent influenza phenotypesis a conse-quence of genetic changes altering the function of individual proteinsortheirfunctionalcompatibility.
Geneticchangesintheviralsurfaceproteinscanimpactonvirus binding,entry,assembly,release,inductionofthehost’simmune responseandantiviraldrugresistance.
Withintheinternalproteins,geneticchangescanimpacton vir-ulenceandinfectivityiftheyaltertheabilityofthevirustoreplicate orinducecellularapoptosis.1Theviralreplicationprocessis
multi-genicandinteractive,involvingthereplicationcomplexandthe structuralandfunctionallyassociatedM1andNS2/NEPproteins.1,2
Thegene segmentscoding for theproteinsinvolvedin replica-tion,codeforadditionalproteinsassociatedwiththeinductionof
∗ Correspondingauthor.Tel.:+351217519332.
E-mailaddress:h.rebelo.andrade@insa.min-saude.pt(H.RebelodeAndrade).
cellular apoptosis during viral infection. The (+1)ORF of PB1 encodesPB1-F2withpro-apoptoticfunctioninthehostcellsand the NS gene encodes NS1 protein associated with the activa-tion of anti-apoptotic mechanisms, permitting viral replication tooccur.3–5Thecassetteofinternalgenesthereforecontrolsthe
major processes by which viral fitness is determined: replica-tionandapoptosis.Anincreasedreplicationrateorstimulationof anti-apoptoticmechanisms,produceshigherviralloadsthatcan overcomehostimmuneresponse,enhanceinfectivityandresultin amoresevereandtransmissibledisease.Themostsevere epidemi-ologicalsituationscausedbyseasonalinfluenzavirusesoccurredin 1947and1951asaconsequenceofgeneticchangeswithin inter-nalproteinsinvolvedinreplicationofA(H1N1)viruses.Thestrains spreadworldwideandcausedexceptionallyhighmortality.6
Monitoringevolutionarytrendsinthegenesencodinginfluenza virusinternalproteins,andprofilingantigenicproteinsand antivi-ral drug susceptibility, are therefore essential to risk assessing influenzavirulence,diseaseepidemiologyandpublichealth impli-cations.
1386-6532/$–seefrontmatter © 2011 Elsevier B.V. All rights reserved. doi:10.1016/j.jcv.2011.11.002
Fig.1.PhylogeneticanalysisoftheaminoacidsequencesofHA1subunitofhemagglutinin(1A)andneuraminidase(1B)genesofinfluenzaA(H1N1)pdm09strainscirculating inPortugal.
2. Objectives
With the aim of risk assessing emergent strains, the main objectivesof thestudywere toprofilegenomic signatures and antiviraldrugsusceptibilityofA(H1N1)pdm09virusesanddiscuss thepotentialputativeroleofmutatedresiduesinhumanhost adap-tationandvirulence.
3. Studydesign
Atotalof110specimensfromlaboratory-confirmedcasesof pandemicinfluenzawerecollectedinPortugalfromJuly2009to January2010.StrainswereisolatedinMDCK-SIAT1cellculture. Antigenicsurface glycoproteins and M1 and M2 proteinswere geneticallycharacterizedandantiviraldrugsusceptibilitywas pro-filed.Geneticcharacterizationofinternalproteinswasperformed onvirusesisolatedfromsubsetsofpregnantwomen,outliersto
NAIsand a 14 years olddeceased patient withnorisk factors. Cyclesequencingwasperformedbasedonadaptationofa proto-colfromCDC,recommendedbyWHO.7Geneticmutationanalysis
andphylogeneticanalysiswereperformedwith LasergeneV.4.05-DNASTAR and MEGA4.0, with comparison to sequences of the A(H1N1)pdm09 2010/2011 vaccine strain (A/California/7/2009) andpublishedreferenceA(H1N1)pdm09strainscirculating world-wide within pandemic and post-pandemic periods. Phenotypic evaluationofantiviraldrugsusceptibilitywasperformedby flu-orescentassaywithMUNANAsubstrateaspreviouslydescribed.8
Minorandmajorphenotypicoutlierswereidentifiedthroughthe establishmentoflowerandupperIC50cutoffsvalues.8Genotypic evaluationofantiviraldrugsusceptibilitytoNAIsandto amanta-dinewasperformedbyanalysingNAandM2proteinsequences, respectively. In the M2 sequence the 5 well-defined molecu-lar markersof resistancetoamantadinewereanalysed: L26F/I; V27A/D;A30T;S31NandG34E.
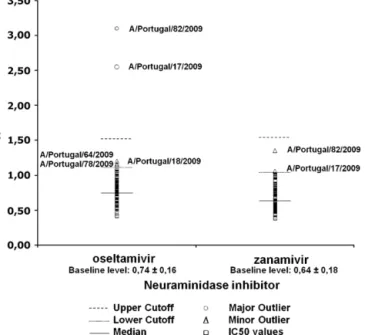
Fig.2.PhenotypicevaluationofA(H1N1)pdm09virussusceptibilitytoNAIsby flu-orescenceassay.
4. Results
Virusisolationwasperformedforall110A(H1N1)pdm09cases. Ofthese,37 weregenetically characterizedfor antigenic glyco-proteinsand for Mprotein. Antiviraldrugsusceptibility profile wasphenotypicallyevaluatedfor103ofthe110isolates. Ninety-eight were furthersequenced for genotypic evaluation of NAIs susceptibilityand96foramantadinesusceptibility.Genetic char-acterizationofinternalproteinsPB2,PB1,PB1-F2,PA,NP,NS1and NS2/NEPwasperformedfor13ofthe37isolates,includingviruses isolatedfromcasesinthesubsetsdescribedabove.
WithintheHA1domainoftheHA,all37strainswereshown tocontainthemutationsP83SandI321V(Fig.1A).Thirty-threeof the37strainshadthesubstitutionS203T,and19alsocontained D222E.Additionalpointmutationswereobserved,includingA73T andS185N inindividualstrains, which arelocatedinthe puta-tiveantigenicsitesCbandSb,respectively.ThesubstitutionK119N whichresultsinthecreationofanN–X–Smotif,wasobservedin 2strains.IntheNAgene,34ofthe37strainsanalysedcontained N248DandV106Isubstitutions(Fig.1B).FifteenstrainshadY155H andtwocontainedI223VintheNAgene.
The genetic characterization of MP of 37 strains identified M128L, E201D and M203I substitutions in individual isolates compared to the A(H1N1)pdm09 2010/2011 vaccine strain MP sequence,locatedwithintheM1codingregion.
Phenotypic evaluation of NAIs susceptibility revealed three minorandtwomajoroutlierstooseltamivir,asshowninFig.2.The threeminoroutliersexhibitedreducedsusceptibilityof approxi-matelytwo-foldandthetwomajoroutliersofapproximately 3-and4-fold,comparedtobaselinelevel.Thetwomajoroutlierswere alsofoundtobeminoroutlierstozanamivir(MjOOsel/MnOZana), withreducedsusceptibilityofapproximatelytwo-foldcomparedto baselinelevel.IntheNAsequenceofthesetwoMjOOselt/MnOZana strains,themutationI223Vwasobserved.Genotypicevaluationof susceptibilitytoamantadinerevealedthatall96strainsanalysed hadanaspargine(N)atposition31intheM2codingregion.
Thegeneticprofileoftheinternalproteinsof13A(H1N1)pdm09 isolatesanalysed revealedsubstitutionsP224S inPA,V100Iand L122QinNPandI123VinNS1,whichwerecommontoall13 iso-lates(highlightedinboldand#,Fig.1),asshowninTable1.PB1-F2
waspresentinthetruncatednon-functionalformof11aminoacids. Table
1 Non-synonymous mutations detected per gene/protein. Strains are ordered according to epidemiologic week of beginning of symptoms. Strain Epidemiological week Gene/protein NS2/NEP Observations PB2 PB1 PA NP NS1 A/Portugal/17/2009 30/2009 V480I – P224S V100I; L122Q I123V – MjO Osel; MnO Zana A/Portugal/82/2009 32/2009 V480I; D680E – P224S V100I; L122Q; K400R I123V – MjO Osel; MnO Zana A/Portugal/2/2009 34/2009 – – P224S V100I; L122Q I123V; F103C – n.o. A/Portugal/7/2009 34/2009 – S633G P224S V100I; L122Q I123V – Pregnancy A/Portugal/8/2009 39/2009 R54K; I463M – P224S; D347N; M561I V100I; L122Q I123V – Pregnancy A/Portugal/13/2009 39/2009 R54K S384L P224S; D347N; C95G; N409S V100I; L122Q I123V S93G Pregnancy A/Portugal/9/2009 43/2009 R54K; I354V S384L P224S; D347N V100I; L122Q I123V; A155V; R211K D54N Pregnancy A/Portugal/10/2009 44/2009 R54K S384L P224S; D347N V100I; L122Q I123V; A155V; R211K D54N; G22E Pregnancy A/Portugal/11/2009 44/2009 R54K; E249G S384L P224S; D347N V100I; L122Q I123V; A155V; R211K D54N Pregnancy A/Portugal/18/2009 45/2009 – – P224S; S296N V100I; L122Q I123V; P114H – MnO Osel; ICU A/Portugal/12/2009 49/2009 R54K – P224S; S364N; N614S V100I; L122Q I123V – Deceased A/Portugal/6/2009 50/2009 E191K – P224S; Q193K; P400L V100I; L122Q I123V – Children A/Portugal/1/2010 04/2010 K340N S678N P224S; I94L; N675H V100I; L122Q; V217I; V313I I123V; A155V – Pregnancy MjO, major outlier; MnO, minor outlier; Osel, oseltamivir; Zana, zanamivir; ICU, intensive care unit; n.o., no observations.
Viruses isolated from the subset of pregnant women, were
observedtohaveR54KsubstitutioninPB2in6cases,5ofwhich
alsocontainedD347NinPA.Ofthese,4alsocontainedS384LinPB1
andasmallclusterof3isolateswascharacterizedbytheadditional
substitutionsA155VandR211KinNS1,andD54NinNS2/NEP.
ThemutationV480IwasfoundonlyinthePB2sequenceofthe
twostrainsexhibitingareducedsusceptibilityprofiletobothNAIs
(MjOOselt/MnOZana).
5. Discussion
A(H1N1)pdm09virusescharacterizedbytheHA substitution
S203T and both N248D and V106I in NA, were found to be
thedominantcirculatingstrainthroughouttheepidemicperiod
(2009–2010)inPortugal.Previousstudieshavealsoreported
dom-inantcirculationofthisstrainworldwideandsuggestedthisisa
consequenceofenhancedviralfitness.9,10
Within HA1, there was no evidence of evolutionary trends, in agreement with the genetic and antigenic homogeneity of A(H1N1)pdm09.11 The location of amino acid substitutions
observedinHA1however,suggeststhesecouldresultin pheno-typicchanges;theD222Esubstitutionislocatedwithinaloopof thereceptor-bindingsite,A73TandS185Nwithintheputative anti-genicsitesCbandSb,respectively,andK119Nhasbeenreportedto resultincreationofapotentialN-glycosylationsite.9,12Correlation
ofthesemutationswithparticularphenotypes,bindingspecificity orclinicaloutcomeofinfectionhowever,needsfurther investiga-tion.
In the internal genes, the V100I and L122Q substitutions observedinNP,I123VinNS1andP224S inPAcharacterizethe geneticprofileofallisolatesanalysed,reflectingthedominant cir-culationofthisA(H1N1)pdm09geneticvariantinPortugalwithin theperiodanalysed.Residues100and122inNParelocatedwithin thebodydomainoftheproteinandarethoughttobeinvolvedin PB2andPB1–NPinteraction,whichiscrucialforRNAreplication. TheaminoacidsubstitutionV100Iwasassociatedwiththe rais-ingofthepandemicalertfromphase4to6,andisahumanto aviansignaturechangeknowntotranslateintoenhancedviral fit-nessprobablybyincreasingviraltransmissibilityorinfectivity.10,13
I123VinNS1islocatedintheeffectordomain,afunctionalregion regulatingcellularapoptosis,andthereforesuspectedtoplayarole inadaptationtothehumanhostandtoincreasevirulence.10P224S
inPAislocatedintheN-terminaldomain,whichhasendonuclease activity.Althoughnophenotypicoutcomeforthis mutationhas beenestablishedtodate,endonucleaseactivityiscriticalfor ini-tiatingtranscriptionandtherefore,geneticchangesinthisregion mightbeexpectedtoalterreplicationkinetics.14Althoughthe
phe-notypicoutcomeofL122Qhasnotbeenestablished,thismutation hasbeenpreviously reported asa virulence factor in H5N1 by greatlyincreasingreplication.15
Inthevirusesfrompregnantwomen,inadditiontothe muta-tionscommontoallisolatesanalysed,auniqueprofileof6genetic signaturesinthefunctionaldomainsofinternalproteinsdefineda specificcluster.Consideringtheirgeneticlocation,theypresenta highpotentialfortranslatingintophenotypicchangesinfitness and virulence. BothR54K in PB2and D347N in PAare located withinfunctionaldomainsforbindingPB1.7,18Sincethefunction
ofthepolymeraseproteinsdepends ontheformation ofa het-erodimerforoptimalviralRNAreplicationandtranscription,these substitutionstogetherwithP224SinPA,mayhaveimpactedon polymerasesubunitbindingtoPB1andhavehadanunpredictable effectonreplicationinthesestrains.16,17WithinNS1,thecluster
isdefinedbyA155VandR211Ksubstitutions,bothlocatedwithin theC-terminaloftheeffectordomain,asisI123V,whichhas pre-viouslybeenfoundtoregulatecellularapoptosis.5,18Replication
andapoptosisarethemajormechanismsthatdefineviralfitness. Theoccurrenceofdifferentpointmutationsasoccurredinstrainsin thiscluster,couldbecompensatoryorhavehadacumulativeeffect leadingtoadifferentphenotype.Thesemutationswillcontinueto bemonitoredinnon-pregnantpatientstoevaluatewhetherthey aresubsetexclusive,resultingfromenhancedfunctionofindividual proteinsorincreasingfunctionalcompatibilityofproteins.
Thevirusisolatefromthedeceasedpatientcontaineda distinc-tivesetofmutationswhichcouldhavecontributedtothismore severeclinicaloutcome. InadditiontoN248DandV106IinNA, S203TinHA,V100IinNPandI123VinNS1,thisisolatecontained L122QinNP,S364NandN614SinPA,andR54KinPB2,alllocated inPB1bindingdomainsoftherespectiveproteins.17Adirect
asso-ciationofPB1bindingtotheotherpolymerasesubunitsandto NPwithamorevirulent phenotypehasnotbeenestablishedto date.However,theformationofPB2–PB1–PAheterodimerandthe bindingofPB2andPB1–NPintoribonucleoproteinsbothdetermine vRNAtranscriptionandcRNAsynthesisforgenomereplicationand therefore,havethepotentialtogreatlyaffecttheviralreplication cycle.17
WithinthesubsetofoutlierstoNAIs,thetwoMjOOsel/MnOZana weredefinedbyI223VinNAandV480IinPB2.Aminoacid substi-tutionsatposition223inNAhavepreviouslybeenshowntoaffect virussusceptibilitytobothoseltamivirandzanamivir, inrecent A(H1N1)pdm09andseasonalviruses.19,20Residue223islocated
intheframeworkoftheNAactivesite,andthusinteractswiththe catalyticresiduestowhichantiviraldrugsbind.21 The
contribu-tionofV480IinPB2tothereducedsusceptibilityprofileobserved is unclear. In addition, nophenotypic impact was observed in A(H1N1)pdm09 viruses containing NA mutation Y155H, previ-ously observed in seasonal A(H1N1) viruses resistant to both NAIs.22
Correlating specific mutations with virulent phenotypes in thehumanhostis notstraightforward.Animal modelsdifferin theirimmune response to infectionand multipleother factors interferewith clinicaland epidemiology outcomes ofinfection. Also, thegeneticbackground of specificmutationsis divergent amonginfluenzastrainsandcanhavedifferentphenotypic out-comes. Several mutations have however, been recognized as virulencemarkers.Althoughsomemechanismsremainunclear,the modificationofgenometranscription,replication,apoptosisand antigenicityaredeterminantsofvirulence.Additionallytoknown molecularmarkersofvirulence,wehaveplacedselectedresidues withinfunctionaldomainsofinternalproteinsthatmayenhance viralfitnessorincreasevirulenceundergenomicsurveillance.
Funding
Calouste Gulbenkian Foundation (Fundac¸ão Calouste Gul-benkian) research grant funded the experimental work and the Portuguese Science and Technology Foundation the doc-toral grants (SFRH/BD/65211/2009; SFRH/BD/62676/2009; SFRH/BD/48532/2008).
Competinginterest Nonedeclared.
Ethicalapproval
Nohumansubjectswereusedinthisresearchstudy.Specimens wereaccessedthroughanonymousbanks,reviewedandexemptfor specificissuedocumentfromtheEthicCommissionoftheNational InstituteofHealth,Lisbon,Portugal.
TheauthorswouldliketothankDr.JoannaEllisfromtheHealth ProtectionAgencyinLondonfortherevisionofthemanuscript. References
1. BaslerC, Aguilar P.Progress in identifyingvirulencedeterminants ofthe 1918H1N1andtheSoutheastAsianH5N1influenzaAviruses.AntiviralRes 2008;79:166–78.
2.ChenL,DavisTC,ZhouH,CoxNJ,DonisRO.Geneticcompatibilityand viru-lenceofreassortantsderivedfromcontemporaryavianH5N1andhumanH3N2 influenzaAviruses.PLoSPathogens2008;4(5):e1000072.
3. HaiR,SchmolkeM,VargaZ,ManicassamyB,WangTT,BelserJA,etal.PB1-F2 expressionbythe2009pandemicH1N1influenzavirushasminimalimpacton virulenceinanimalmodels.JVirol2010;84(9):4442–50.
4.HaleBG,RandallRE,OrtínJ,JacksonD.ThemultifunctionalNS1proteinof influenzaAviruses.JGenVirol2008;89(Pt10):2359–76.
5. ZhirnovOP,KonakovaTE,WolffT,KlenkHD.NS1proteinofinfluenzaAvirus down-regulatesapoptosis.JVirol2002;76(4):1617–25.
6.NelsonMI,ViboudC,SimonsenL,BennettRT,GriesemerSB,StGeorgeK,etal. MultiplereassortmenteventsintheevolutionaryhistoryofH1N1influenzaA virussince1918.PLoSPathogens2008;4(2):e1000012.
7.WorldHealthOrganization.Sequencingprimersandprotocol;2009.Availableat:
http://www.who.int/csr/resources/publications/swineflu/sequencingprimers/ en/index.html(accessed15.10.10).
8. CorreiaV,deAndradeHR,SantosLA,LackenbyA,ZambonM.Antiviraldrug profileofseasonalinfluenzavirusescirculatinginPortugalfrom2004/2005to 2008/2009winterseasons.AntiviralRes2010;86:128–36.
9.IgarashiM,ItoK,YoshidaR,TomabechiD,KidaH,TakadaA.Predictingthe anti-genicstructureofthepandemic(H1N1)2009influenzavirushemagglutinin. PLoSOne2010;5(1):e8553.
10.PanC,CheungB,TanS,LiC,LiL,LiuS,etal.Genomicsignatureandmutationtrend analysisofpandemic(H1N1)2009influenzaAvirus.PLoSOne2010;5(3):e9549.
use inthe 2011southern hemisphereinfluenza season; 2010. Available at:
http://www.who.int/csr/disease/influenza/201009Recommendation.pdf
(accessed16.12.10).
12.KilanderA,RykkvinR,DudmanSG,HungnesO.Observedassociationbetween theHA1mutation D222G inthe2009 pandemicinfluenzaA(H1N1) virus and severeclinical outcome,Norway 2009–2010. EuroSurveill 2010;15(9), pii=19498.
13.WuR,ZhangH,YangK,LiangW,XiongZ,LiuZ,etal.Multipleaminoacid sub-stitutionsareinvolvedintheadaptationofH9N2avianinfluenzavirustomice. VetMicrobiol2009;138:85–91.
14. LiuY,LouZ,BartlamM,RaoZ.Structure–functionstudiesoftheinfluenzavirus RNApolymerasePAsubunit.SciChinaSerCLifeSci2009;52(5):450–8. 15. TadaT,SuzukiK,SakuraiY,KuboM,OkadaH,ItohT,etal.NPBodyDomain
andPB2ContributetoIncreasedVirulenceofH5N1HighlyPathogenicAvian InfluenzaVirusesinChickens.JVirol2011;85(4):1834–46.
16.GuuTS,DongL,Wittung-StafshedeP,TaoYJ.Mappingthedomainstructureof theinfluenzaAviruspolymeraseacidicprotein(PA)anditsinteractionwiththe basicprotein1(PB1)subunit.Virology2008;379:135–42.
17. PerezDR,DonisRO.FunctionalanalysisofPAbindingbyinfluenzaAvirusPB1: effectsonpolymeraseactivityandviralinfectivity.JVirol2001;75(17):8127–36. 18.SalahuddinP,KhanAU.StructuralandfunctionalanalysisofNS1andNS2
Pro-teinsofH1N1subtype.GenProtBioinform2010;8(3):190–9.
19.HaydenFG,deJongMD.Emerginginfluenzaantiviralresistancethreats.JInfect Dis2011;203(1):6–10.
20.DeydeVM,SheuTG,TrujilloAA,Okomo-AdhiamboM,GartenR,KlimovAI, etal.Detectionofmolecularmarkersofdrugresistancein2009pandemic influenzaA(H1N1)virusesbypyrosequencing.AntimicrobAgentsChemother 2010;54(3):1102–10.
21.YenHL,HoffmannE,TaylorG,ScholtissekC,MontoAS,WebsterRG,etal. Impor-tanceofneuraminidaseactive-siteresiduestotheneuraminidaseinhibitor resistanceofinfluenzaviruses.JVirol2006;80(17):8787–95.
22.MontoAS,McKimm-BreschkinJL,MackenC,HampsonAW,HayA,Klimov A,etal.Detectionofinfluenzavirusesresistanttoneuraminidaseinhibitors inglobalsurveillanceduringthefirst3yearsoftheiruse.AntimicrobAgents Chemother2006;50(7):2395–402.