ISSN 1678-4375 (Online)
Genetic variability of populations of the white-eared opossum,
Didelphis albiventris
Lund 1840 (Didelphimorphia; Didelphidae)
in Brazil
D. C. Nascimento
a,b, B. A. T. P. Campos
c, E. C. Fraga
a,b,cand M. C. Barros
a,b,c*
aPrograma de Mestrado em Ciência Animal – CCMA, Centro de Ciências Agrárias – CCA, Universidade Estadual doMaranhão – UEMA, Cidade Universitária Paulo VI, CP 9, São Luís, MA, Brasil
bCentro de Estudos Superiores de Caxias – CESC, Laboratório de Genética e Biologia Molecular – GENBIMOL,
Universidade Estadual do Maranhão – UEMA, Praça Duque de Caxias, s/n, Morro do Alecrim, CEP 65604-380, Caxias, MA, Brasil
cPrograma de Pós-graduação em Biodiversidade, Ambiente e Saúde – PPGBAS, Centro de Estudos Superiores de
Caxias – CESC, Universidade Estadual do Maranhão – UEMA, Praça Duque de Caxias, s/n, Morro do Alecrim, CEP 65604-380, Caxias, MA, Brasil
*e-mail: mbdene@yahoo.com.br
Received: September 01, 2017 – Accepted: February 10, 2018 – Distributed: November 30, 2019
(With 4 figures)
Abstract
Didelphis albiventris are found throughout Northeast and Central Brazil to central-southern Uruguay and it was subject of few studies in a population level. Given this, the present study investigated the genetic variability of the species using the mitochondrial molecular marker cytochrome oxidase c subunit I. We analyzed samples from the different
biomes within three Brazilian regions: Northeast (Caatinga, Cerrado, and Atlantic Forest), Southeast (Cerrado, Atlantic
Forest, Cerrado/Atlantic Forest, and Cerrado/Caatinga ecotones) and South (Pampa and Atlantic Forest). Software BAPs retrieved five distinct demes: dm 1, dm 2, and dm 5 that occurs in South, Northeast and Southeast regions respectively
and the dm 3 and dm 4 are wide distributed in Northeast and Southeast. Population analysis performed with AMOVA, haplotype network and Mantel test estimated the veracity of the demes. The FST shows structuring for the five demes, with dm 1 (South region) isolated from the others, however the other analysis showed the Northeast/Southeast demes (dm 2-5) united, diagnosing gene flow between them, mainly at the transitional zones, in areas as far away as areas with similar latitude interval (Southeast vs South) that was not detected gene flow. In the haplotype network, the mutational
steps was conclusive in split dm1 from dm 2-5 with 15 mutational steps and the Mantel test was moderated, which
is explained by genetic similarity despite the great geographic distances (Northeast/Southeast). Thus, our analysis recognized two different lineages (South and Northeast/Southeast) and indicate that the biomes were not decisive in their isolation. The sharing of demes at the transitional zones and in areas with high latitudinal intervals reflects a recent
ancestral polymorphism for D. albiventris. The plasticity in the occupation of the space by this species contributes in its wide dispersion capability, that is, geographical distribution. Our results revealed important implications for the management of D. albiventris in these transitional zones areas where demes were shared.
Keywords: marsupials, COI, population diversity, geographic lineages.
Variabilidade genética de populações do gambá de orelha branca,
Didelphis albiventris
Lund 1840 (Didelphimorphia; Didelphidae) no Brasil
Resumo
Didelphis albiventris é encontrada em todo o Nordeste e região central do Brasil até o centro-sul do Uruguai e foi alvo de poucos estudos em nível populacional. Dessa forma, o presente estudo, investiga a variabilidade genética da espécie usando o marcador molecular citocromo c oxidase subunidade I. Analisou-se amostras de diferentes biomas de três regiões brasileiras: Nordeste (Caatinga, Cerrado e Floresta Atlântica), Sudeste (Cerrado, Floresta Atlântica,
ecótonos Cerrado/Floresta Atlântica e Cerrado/Caatinga) e Sul (Pampa e Floresta Atlântica). O software BAPs recuperou cinco demes distintos: dm 1, dm 2 e dm 5, que ocorrem nas regiões Sul, Nordeste e Sudeste, respectivamente, e os dm 3 e dm 4, que são amplamente distribuído no Nordeste e Sudeste. Análises populacionais realizadas com AMOVA, rede de haplótipo e teste de Mantel estimaram a veracidade das demes. O FST mostrou estruturação para as cinco demes,
1. Introduction
The marsupials of the genus Didelphis Linnaeus, 1758 are a small non-volant mammals that are an essential component of natural communities and good indicators of environmental changes and its degree of conservation (Conde y Vera and Rocha, 2006; Castro, 2012; Quintela et al., 2013). In particular, populations of the white-eared opossum (Didelphis albiventris Lund, 1840) tends to increase in density in response to environmental disturbance (Cáceres et al., 2008), with associated implications for public health given that this species is a natural reservoir for a number of human diseases such as Chagas disease and leishmaniasis (Sherlock et al., 1984; Pinho et al., 2000; De Luca et al., 2003; Bern et al., 2011; Zetun et al., 2014).
Due to its generalist habit, D. albiventris is highly flexible in the use of space, exploiting a variety of vertical strata and occupying large home ranges (Almeida et al., 2008; Sanches et al., 2012). The species is widely distributed in deciduous forests, occurring from northeastern and central Brazil to central and southern Uruguay, as well as in Paraguay, Argentina and Bolivia (Cerqueira and Tribe, 2008). In Brazil there are records in Caatinga, Cerrado, Pantanal, Pampas habitats and transitional zones between these biomes (Cerqueira and Tribe, 2008; Paglia et al., 2012).
Despite the genus Didelphis is common and ecologically successful (Durant, 2001), little is known about the relationships among of D. albiventris populations, with only two studies available: Sousa et al. (2012) and
Rocha et al. (2015). Both works revealed some degree of genetic structuring, the former studied the mitochondrial gene cytochrome c oxidase subunit I (COI) of populations
from southern and southeastern Brazil, and the other the mitochondrial gene cytochrome b from few localities in central Brazil.
Mitochondrial DNA have revealed degree of population structure for many species, being the most used molecular marker for tracing the geographic distribution of genealogical lineages (Avise et al., 1987). Males and females of the D. albiventris present similar patterns of dispersal
(Sanches et al., 2012), and the sex of the specimen should be irrelevant to the selection of the molecular marker, consequently the matrilineal inheritance of the mitochondria should not interfere in the analyses. Notably, mitochondrial genes have been used successfully in population-level and
phylogenetic studies of small mammals (Carvalho et al., 2011; Agrizzi et al., 2012; Sousa et al., 2012; Muller et al., 2013; Rocha et al., 2015). In particular, the COI gene has
been used as a DNA barcode for the identification of a number of different vertebrate groups (Hebert et al., 2003),
and has contributed important findings on D. albiventris
populations (Sousa et al., 2012).
Ecological studies use population and genetic data in attempt to understand the biology of a species. With the scarcity of population genetic information throughout its distribution, especially for the inhabitants of Cerrado and Caatinga, the present work investigated how D. albiventris
populations are distributed in Brazil using the COI as a molecular marker: 1. The species possesses populations structured in the Northeast/South axis? 2. There is more than one population per Biome? 3. If yes, they are shared between Biomes or are exclusive for one Biome? Ecotone
areas has a different role in the population structure of the
species? 4. Is there natural selection or are the populations selectively neutral? 5. In attempt to answer these questions we conducted genetic variability study of populations of the white-eared opossum, D. albiventris.
2. Material and Methods
2.1. Description of the samples
Samples of D. albiventris were obtained from the Northeast, Southeast and South regions of Brazil, between latitudes 04°05’ S and 30º01’ S (Figure 1). A total of 52 samples were obtained in the Northeast region (Paraíba/PB, Sergipe/SE, Pernambuco/PE, Ceará/CE,
Piauí/PI and Maranhão/MA), from the Atlantic Forest,
Caatinga and Cerrado biomes. Eighty four samples from
Southeast (Minas Gerais/MG) from the Atlantic Forest
and Cerrado biomes, and the Cerrado/Atlantic Forest and Cerrado/Caatinga ecotones. Six samples from South
(Rio Grande do Sul/RS) corresponding to the Pampas
and Atlantic Forest (Figure 1; Supplementary Material). The samples from the Northeast were obtained in the Universidade Estadual do Maranhão, for which the collecting data are available in Costa et al. (2012), and
from collaborations with the scientific collections of other
Brazilian research institutions, such as Universidade Federal da Paraíba (UFPB), Universidade Federal de Sergipe (UFS), Universidade Federal do Piauí (UFPI),
and Museu Paraense Emílio Goeldi (MPEG). All samples
áreas com similar intervalo de latitude (Sudeste e Sul), onde não foram detectado fluxo gênico. Na rede de haplótipo,
os passos mutacionais foram conclusivos em separar dm 1 do dm 2-5 com 15 passos mutacionais, e o teste de Mantel
foi moderado, o que é explicado pela similaridade genética apesar da grande distância geográfica (Nordeste/Sudeste). Assim, duas linhagens diferentes (Sul e Sudeste/Nordeste) foram encontradas, indicando que os biomas não foram
decisivos em seus isolamentos. Os compartilhamentos das demes, em zonas de transição e em áreas com elevados
intervalos de latitude, refletem um polimorfismo ancestral recente para D. albiventris. A plasticidade na ocupação do espaço por esta espécie contribui em sua ampla capacidade de dispersão, ou seja, distribuição geográfica. Nossos
resultados revelam importantes implicações para o manejo de D. albiventris nessas áreas de zonas de transição, onde
as demes são compartilhadas.
from Southeast and South were obtained at GenBank from
Sousa et al. (2012).
2.2. DNA extraction, amplification and sequencing DNA was extracted from muscle, liver, and ear tissue using the Phenol-Chloroform protocol of Sambrook and
Russel (2001). We amplified and sequenced a fragment
of the mtDNA COI gene using the primers LCO1490 and HCO2198 (Folmer et al., 1994). The COI gene was
amplified in 25 μl reactions of PCR, consisting of 4.0 μl of dNTP (1.25 nM), 2.5 μl of buffer (10X), 0.5 μl of MgCl2
(25 nM), 0.25 μl of each primer (200 ng/μl), 0.2 μl of Taq DNA Polymerase 10 x 250 μl (GE Healthcare), and 1 μl of template DNA (100 ng/μl), with the final volume being completed by purified water. The samples were amplified in a Veriti 96 well thermal cycler (Applied Biosystems) with an initial denaturation (94ºC for 3 min), 40 cycles of denaturation (94ºC for 45 s), annealing (48ºC for 45 s), and extension (72ºC for 1 min and 30 s), and a final extension
at 72ºC for 3 min. We used the BigDye Terminator v3.1
Cycle Sequencing Kit (Applied Biosystems) for sequencing
reactions. The sequences were obtained using an ABI 3500 automatic DNA sequencer with associated Sequencing
Analysis software (Applied Biosystems).
2.3. Data analysis
The sequences were aligned using Clustal W (Thompson et al., 1994) in BioEdit v7.0.5.2 (Hall, 1999). The analysis of the variable sites was performed in Mega v6.06 (Tamura et al., 2013). Haplotype and nucleotide diversity were estimated in DnaSP v4.9 (Rozas et al., 2003),
and the groups used for the population analyses were defined
using the BAPs software (Corander et al., 2008). In this method, the number of populations is treated as unknown parameter and is directly inferred from the data set without
defining a prior estimate, a dendogram were generated
using Unweighted Pair Group Method using Arithmetic
averages - UPGMA. The populations identified by this
Figure 1. Localities from which the samples were obtained for the present study. The demes were defined according to the
BAPs software and are represented by geometric figures with numbers (designated in Supplementary Material); Biomes are
software were used to structure the subsequent population analyses. The haplotype networks were produced in NetWork v4.6.1.2 (Bandelt et al., 1999). An Analysis of Molecular
Variance (AMOVA) was performed in Arlequin v3.5.2.1
(Excoffier and Lischer, 2010) to obtain the index of allelic
fixation (FST). The neutrality tests D (Tajima, 1989) and Fu’s FS (Fu, 1997) were run in Arlequin v3.5.2.1 (Excoffier and Lischer, 2010). The Mantel test, which relates genetic distance to geographic distance, was run in the Alleles in Space software with 10,000 replications (Miller, 2005).
3. Results
3.1. Molecular characterization and neutrality tests The study included a total of 142 sequences for the mitochondrial COI gene of D. albiventris, made up of a fragment of 655 bps, of which 624 bps were conserved,
31 were variable, and 21 were informative for parsimony analyses, corresponding to 19 haplotypes.
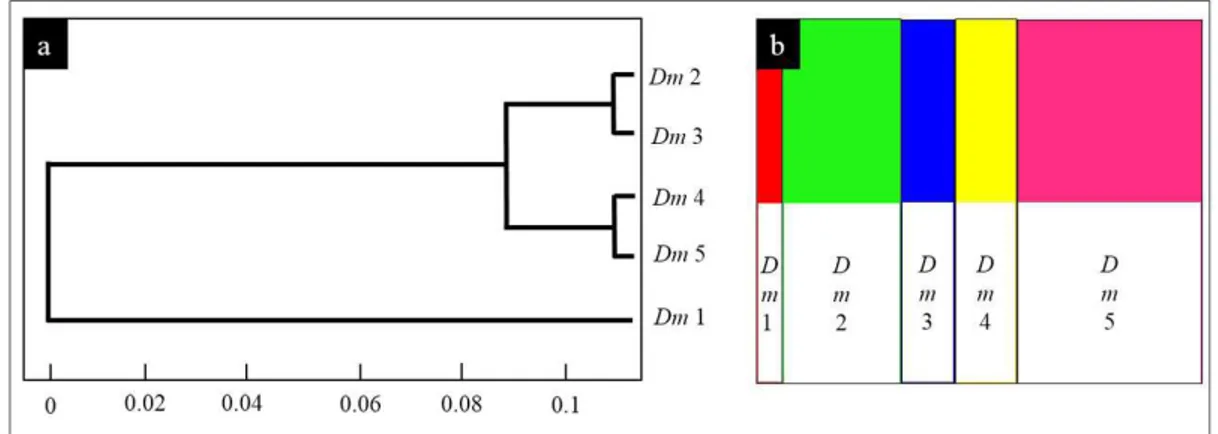
The sequences plotted in the BAPs software (Figure 2)
indicated the existence of five Brazilian demes (dm), as
can be seen in the haplotype network and in the map (Figure 1 e 3). The dm 1 occurs in the Pampas and Atlantic
Forest biomes (South region; H1 and H2), dm 2 in the
Caatinga and Atlantic Forest biomes (Northeast region;
H3-H8) and dm 5 Cerrado, Caatinga, Atlantic Forest
and ecotones (Southeast regions; H16-H19). The dm 3 (H 9-H13) and dm 4 (H14 and H15) are more widely
distributed, being found simultaneously in the Cerrado and Caatinga (Northeast region) and Cerrado, Caatinga,
Atlantic Forest (Southeast region). We can observe that some localities present multiple demes: Xacriabá Amerindian
Reservations/MG that contains dm 3, dm 4 and dm 5; Caxias/MA dm 3 and dm 4, Belo Horizonte/MG and Nova Lima/MG dm 4 and dm 5 (Table 1).
Figure 2. Didelphis albiventris demes defined through the BAPs software for the COI gene. (a) UPGMA tree using the Nei
model. (b) Mixture among the demes obtained, the size of the color bars is proportional to the number of specimens in the
populations.
Figure 3. Haplotype network for the COI gene in the D. albiventris demes. The numbers represents the mutational sites between
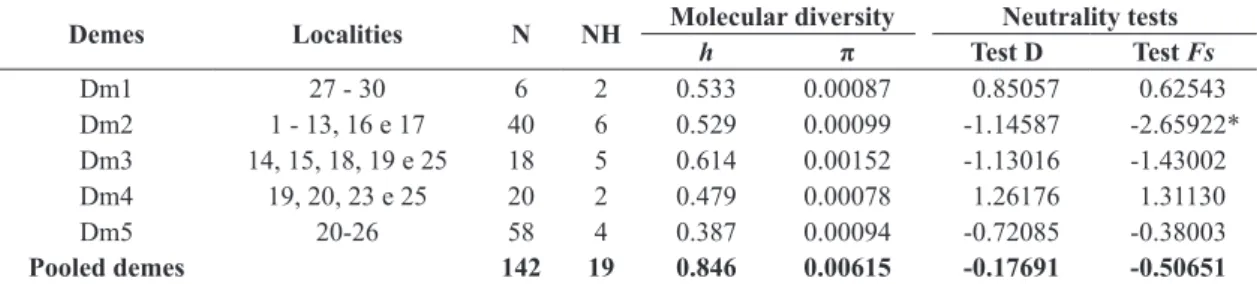
The total haplotypic diversity (h) was h = 0.846, and varied from h = 0.387 for dm 5 to h = 0.614 for dm 3,
while total nucleotide diversity (π) was π = 0.00615, and varied from π = 0.00078 for dm 4 to π = 0.00152 for dm
3 (Table 1). No significant p values were found in any deme for the D test (Tajima, 1989), and a significant value for Fu’s FS (Fu, 1997) was only recorded for dm 2,
indicating that only this deme is influenced by demographic
parameters (Table 1).
3.2. Genetic differentiation among demes
AMOVA analyses based on the five demes retrieved
by the BAPs analysis (Table 1) indicated that 87.47% of
the variation was due to differences among the demes, and 12.53% within the demes, with a highly significant
(p = 0.00000) FST value of 0.87 (Table 2). The pairwise
FST values were higher, ranging from 0.7, for dm 2 vs. dm 3, to 0.97 for dm 4 vs. dm 1 (Table 3).
The results of the Mantel test for the five demes
indicated a moderate degree of correlation between genetic and geographic distances (r = 0.54; p = 0.00001; Figure 4).
When dm 1 (r = -0.31; p = 0.19), dm 2 (r= -0.13; p = 0.79), and dm 5 (r= -0.04; p = 0.61) were analyzed separately the correlation were low, although dm 3 (r = 0.90; p = 0.001) and dm 4 (r = 0.89; p = 0.001) returned high correlations.
4. Discussion
Due to the ecological importance of D. albiventris, the investigation of the dynamics of dispersion of its populations through a genetic approach will improve the knowledge
and contribute to the establishment of management and conservation measures along its geographical distribution.
Were analyzed the largest sample for this specie to date, and high levels of haplotype diversity (h) and nucleotide
diversity (π) were found. Similar results were retrieved
in Sousa et al. (2012); h = 0.7235; π = 0.0065) for COI and Rocha et al. (2015) for Cytochrome b (h = 0.876;
π = 0.0062), reflecting the high genetic diversity of this
species. According to Grant and Bower (1998) high values of h and π combined (above 0.5 and 0.005 respectively)
Table 1. Genetic diversity and the results of the neutrality tests for the COI gene in the D. albiventris demes analyzed.
Demes Localities N NH Molecular diversity Neutrality tests
h π Test D Test Fs
Dm1 27 - 30 6 2 0.533 0.00087 0.85057 0.62543
Dm2 1 - 13, 16 e 17 40 6 0.529 0.00099 -1.14587 -2.65922*
Dm3 14, 15, 18, 19 e 25 18 5 0.614 0.00152 -1.13016 -1.43002
Dm4 19, 20, 23 e 25 20 2 0.479 0.00078 1.26176 1.31130
Dm5 20-26 58 4 0.387 0.00094 -0.72085 -0.38003
Pooled demes 142 19 0.846 0.00615 -0.17691 -0.50651
N: Sample size; NH: Number of haplotypes; h: Haplotype diversity; π: Nucleotide diversity; (*) = p < 0.05.
Table 3. Pairwise FST values for the COI gene for five demes of D. albiventris.
Deme Dm 1 Dm 2 Dm 3 Dm 4 Dm 5
Dm 1 0.00000
Dm 2 0.96492 0.00000
Dm 3 0.94811 0.71389 0.00000
Dm 4 0.97321 0.87873 0.85555 0.00000
Dm 5 0.96605 0.87873 0.81081 0.72012 0.00000
P = 0.0000.
Table 2. Results of the AMOVA for the COI gene for the five Didelphis albiventris demes retrieved from BAPs.
Source of variation Variance component % Variation FST p*
Among demes 2.18911 87.47 0.87465 0.0000
Within demes 0.31372 12.53
*Values of P, calculated randomly through 1023 permutations.
Figure 4. Results of the Mantel test for the correlation
between genetic (y) and geographic (x) distances in the COI
reflects long evolutionary history. In the present study our
results shows the haplotypes H11, H14 and H16 widely geographic distributed, suggesting long dispersion time and consequently long evolutionary history, corroborating
Grant and Bower (1998).
By randomly treating allelic frequencies of genetic marker and genetic divergence between groups, the BAPS
software retrieved five demes, which are divided between
the three regions analyzed, with overlap for demes 3 and 4 in the Northeast and Southeast Brazil. The BAPS was not unanimous in considering the geographic proximity or the biomes to delimit the demes, consequently other population analyzes were carried out to verify if they correspond to
different lineages.
In order to identify if there is selective neutrality, tests
were performed for the five demes. The results of Tajima’s D were not significant and Fu’s Fs was only significant
for dm 2, which contains haplotypes from the Northeast.
Significant results for these tests indicate the presence of
natural selection or demographic processes among the variants of a nucleotide sequence in a population. Dm 2 is
distributed over a wide area, with different kind of biomes
which could be interfering in the selective neutrality (Fu and Li, 1993; Nielsen 2001). No significant results were obtained for these same tests by Sousa et al. (2012) and Rocha et al. (2015), which indicates that this species is relatively constant. It is important to mention that these authors did not study northeastern populations.
The results of the AMOVA for the five demes indicate
structuring (FST = 0.87). This structuring is evident in the
case of dm 1, which has pairwise FST values of 0.95 to 0.97,
indicating possible processes of allele fixation and genetic
isolation. Sousa et al. (2012) also found evidence of population structuring (FST> 91%) in the Southeast and
South populations.
The remaining demes analyzed in the AMOVA
(dms 2, 3, 4 and 5) had also high pairwise FST values
(0.71 to 0.88). Despite the high FST, the same demes are
distributed in different localities, for example: dm 3 are in Caxias/MA and Xacriabá Amerindian Reservations/MG; dm 4 in Caxias/MA, Xacriabá Amerindian Reservations/MG, Belo Horizonte/MG and Nova Lima/MG; dm 5 in Xacriabá
Amerindian Reservations/MG, Belo Horizonte/MG
and Nova Lima/MG. We can also notice that different
demes are in sympatry, for example: dm 3 and dm 4 in
Caxias/MA; dm 3, dm 4 and dm 5 in Xacriabá Amerindian
Reservations/MG; dm 4 and dm 5 in Belo Horizonte/MG and Nova Lima/MG.
These sympatric demes show that FST are considering
different structured population in the same localities.
This condition could be explained by the wide distribution of D. albiventris (Figure 1) and by the highly adaptable and generalist habit of the specie, since it is known that altitude and environment do not interferes in the specie distribution (Aragona and Marinho-Filho, 2009). Thus, although FST shows structuring, the connection between demes allows that Northeastern and Southeastern regions belong to the same widespread lineages.
Studies of didelphids have shown population
structuring among different Brazilian biomes, for example Carvalho et al. (2011) and Caramaschi et al. (2011), found population structuring of Monodelphis domestica between the Cerrado and Caatinga. In the present study this
relationship was not verified, as the demes are not restricted to one specific biome and are dispersed among a number of different biomes. This fact may be associated in differences
in home ranges and in ecological characteristics between M. domestica and D. albiventris that could be possible
directly related with body size. Didelphis albiventris is the largest Didelphidae weighting 500-2,750g while M. domestica weight 80-120g only. This size difference
reflects in greater dispersion capacity (Reis et al., 2006; Wang et al., 2009) of D. albiventris that increases the gene
flow, making population structuring more difficult. In this
study, only dm 1 was isolated from the others, but this fact is not associated with biomes, since that deme occurs in Atlantic Forest and Pampa’s biomes.
When comparing the mutational sites among the demes, the haplotype network also shows two lineages
with different geographic distributions for D. albiventris
in this study: one Southern lineages (dm 1) and the other comprising Northeast/Southeast (demes 2-5) showed shared
haplotypes. These two principal lineages are separated by 15 mutational steps. Northeast and Southeast dm’s are
separated by few mutational steps (maximum of five, see
Figure 3), have distribution overlapping between biomes and therefore should be considered as a single lineage.
The gene flow shown in the haplotype network represents
an ancestral polymorphism, since the shared haplotypes are distributed over long geographic distances, as can be
seen in dm 3 (H11) that occurs in Rio Grande do Piauí/PI
(Cerrado) and Xacriabá Amerindian reservations/MG (Cerrado/Caatinga ecotone) (latitudes 07º 45’ S - 14° 53’ S). But the results of the haplotype network did not reveal
connection between dm 5 (H16 and18) in Piracema/MG (Atlantic Forest) and dm 1 (H1) Machadinho/RS (Atlantic Forest) which has similar latitude interval (latitudes 20º 30’ S - 27º 34’ S) (Figure 1 and 3; Table S1), another indication of the isolation of these two strains.
The Mantel test indicated a moderate degree of correlation between genetic and geographic distances in the
five demes. In this study, with the addition of D. albiventris samples from the Northeast, we obtained a reduction in the correlation obtained by Sousa et al. (2012). This reduction in the correlation reinforces the similarity found between demes 2-5, not diagnosed by these authors, and indicates that the Northeast/Southeast lineage is genetically similar although geographically distant.
Northeast and Southeast are connected. It is important to mention that connection of demes occurs in the transitional areas between biomes, which strengthens the conclusions of Sousa et al. (2012) and Rocha et al. (2015) about the importance of the transition zones for research and planning.
These areas are considered as hotspots of intraspecific
diversity and play an important role in the genetic diversity of species and should be closely investigated (Smith, 1997; Kark et al., 2002).
Our results provide important new insights into the population dynamics of small mammals in Brazil. It has long been assumed that these kind of mammals have low vagility, with geographically structured populations and highly divergent genetic traits (Crisci et al., 2003). The extensive connection between demes of the Northeast and Southeast for D. albiventris may reflect the ecological plasticity of this species, resembling a medium-large mammal, with large home-range.
In the present work we diagnosed two lineages
(Northeast/South axis) for the species D. albiventris and that the biomes do not interfere in the isolation of their
demes. However, as an essential finding, the present study
evidences extensive contact between demes at the transition zones, serving as reference to the conduction of important insights for the development of management actions.
Acknowledgements
We are grateful to Fundação de Amparo à Pesquisa e Desenvolvimento Científico do Maranhão (FAPEMA) for financing this study, and the students of the GENBIMOL
(Genetics and Molecular Biology Laboratory, Caxias,
Maranhão, Brazil) for their invaluable support in the field. This study was financed in part by the Coordenação de
Aperfeiçoamento de Pessoal de Nível Superior -Brasil
(CAPES) - Finance code 001.
References
AGRIZZI, J.A., LOSS, C., FARRO, A.P.C., DUDA, R., COSTA, L.P. and LEITE, Y.L.R., 2012. Molecular diagnosis of atlantic forest mammals using mitochondrial DNA sequences: Didelphid Marsupials. The Open Zoology Journal, vol. 5, no. 1, pp. 2-9. http://dx.doi.org/10.2174/1874336601205010002.
ALMEIDA, A.J., TORQUETTI, C.G. and TALAMONI, A.S., 2008. Use of space by neotropical marsupial Didelphis albiventris
(Didelphimorphia: Didelphidae) in an urban forest fragment.
Revista Brasileira de Zoologia, vol. 25, no. 2, pp. 214-219. http:// dx.doi.org/10.1590/S0101-81752008000200008.
ARAGONA, M. and MARINHO-FILHO, J., 2009. História natural e biologia reprodutiva de marsupiais no Pantanal, Mato Grosso, Brasil. Zoologia, vol. 26, no. 2, pp. 220-230. http://dx.doi. org/10.1590/S1984-46702009000200004.
AVISE, J.C., ARNOLD, J., BALL, R.M., BERMINGHAM, E., LAMB, T., NEIGEL, J.E., REEB, C.A. and SAUNDERS, N.C., 1987. Intraspecific Phylogeography: The mitochondrial DNA bridge between population genetics and systematics. Annual Reviews, vol. 18, pp. 489-522.
BANDELT, H.J., FORSTER, P. and RÖHL, A. 1999. Median-joinng networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, vol. 16, no. 1, pp. 37-48.
BERN, C., KJOS, S., YABSLEY, M.J. and MONTGOMERY, S.P., 2011. Trypanosoma cruzi and Chagas’ Disease in the United States. Clinical Microbiology Reviews, vol. 24, no. 4, pp. 655-681. http://dx.doi.org/10.1128/CMR.00005-11. PMid:21976603.
CÁCERES, N.C., CASELLA, J., VARGAS, C.F., PRATES, L.Z., TOMBINI, A.A.M., GOULART, C.S. and LOPES, W.H., 2008. Distribuição geográfica de pequenos mamíferos não voadores nas bacias dos rios Araguaia e Paraná, região centro-sul do Brasil. Iheringia. Série Zoologia, Porto Alegre, vol. 98, no. 2, pp. 173-180. http://dx.doi.org/10.1590/S0073-47212008000200001.
CARAMASCHI, F.P., NASCIMENTO, F.F., CERQUEIRA, R., and BONVICINO, C.R., 2011. Genetic diversity of wild populations of the grey short-tailed opossum, Monodelphis domestica
(Didelphimorphia: Didelphidae), in Brazilian landscapes. Biological
Journal of the Linnean Society, vol. 104, no. 2, pp. 251-263.
CARVALHO, B.A., OLIVEIRA, L.F.B., LANGGUTH, A., FREYGANG, C.C., FERRAZ, R.S. and MATTEVI, M.S., 2011. Phylogenetic relationships and phylogeographic patterns in Monodelphis (Didelphimorphia: Didelphidae). Journal of Mammalogy, vol. 92, no. 1, pp. 121-133. http://dx.doi. org/10.1644/10-MAMM-A-075.1.
CASTRO, K.C., 2012. Assembleia de pequenos mamíferos não voadores da floresta nacional do Amapá, Amazônia oriental. Macapá: Universidade Federal do Amapá, 85 p. Dissertação de Mestrado em Biodiversidade Tropical.
CERQUEIRA, R. and TRIBE, C.J. 2008 Genus Didelphis. In: A. L. GARDNER, ed. Mammals of South America. 1. Marsupials, xenarthrans, shrews, and bats. Chicago, Illinois: University of Chicago Press, pp. 17-25.
CONDE Y VERA, C.F. and ROCHA, C.F., 2006. Habitat disturbance and small mammal richness and diversity in an Atlantic Rainforest Area in Southeastern Brazil. Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 66, no. 4, pp. 983-990. http://dx.doi.org/10.1590/S1519-69842006000600005. PMid:17299934.
CORANDER, J., MARTTINEN, P., SIRÉN, J. and TANG, J., 2008. Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC Bioinformatics, vol. 9, no. 539, pp. 1-14. http://dx.doi.org/10.1186/1471-2105-9-539. PMid:19087322.
COSTA, J.F., NASCIMENTO, D.C., SANTOS, L.L.L., OLÍMPIO, A.P.M., FRAGA, E.C. and BARROS, M.C. 2012. Pequenos mamíferos não voadores de ocorrência na Área de proteção Ambiental Municipal do Inhamum, Caxias/MA. In: M. C. BARROS, ed. Biodiversidade na área de proteção ambiental municipal do Inhamum. São Luís: UEMA, pp. 117-130. CRISCI, J.V., KATIANAS, L. and POSADAS, P. 2003. Historical Biogeography: An introduction. Massachusetts: Havard University Press, 170 p.
DE LUCA, A.S., VASCONCELOS, H.L. and BARRETT, T.V.,
2003. Distribution of sandflies (diptera: phlebotominae) in forest
remnants and adjacent matrix habitats in Brazilian Amazonia. Brazilian Journal of Biology = Revista Brasileira de Biologia, vol. 63, no. 3, pp. 401-410. http://dx.doi.org/10.1590/S1519-69842003000300006. PMid:14758699.
DURANT, P., 2001. Black-eared Opossum Didelphis marsupialis
EXCOFFIER, L. and LISCHER, H.E.L., 2010. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources, vol. 10, no. 3, pp. 564-567. http://dx.doi.org/10.1111/j.1755-0998.2010.02847.x. PMid:21565059.
FOLMER, O., BLACK, M., HOEH, W., LUTZ, R. and VRIJENHOEK, R., 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, vol. 3, no. 5, pp. 294-299. PMid:7881515.
FU, Y.X. and LI, W.H., 1993. Statistical tests of neutrality of mutations. Genetics, vol. 133, no. 3, pp. 693-709. PMid:8454210. FU, Y.X., 1997. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics, vol. 147, no. 2, pp. 915-925. PMid:9335623.
GRANT, W.S. and BOWER, B.W., 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. The American Genetic Association, vol. 89, pp. 415-426.
HALL, T.A., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series, vol. 41, pp. 95-98.
HEBERT, P.D., RATNASINGHAM, N.S. and DEWAARD, J.R., 2003. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings. Biological Sciences, vol. 270, suppl. 1, pp. S96-S99. http://dx.doi. org/10.1098/rsbl.2003.0025. PMid:12952648.
KARK S., MUKERJI, T., SAFRIEL, U.N., NOY-MEIR, I., NISSANI, R. and DARVASI, A., 2002. Peak morphological diversity in an ecotone unveiled in the chukar partridge by a
novel Estimator in a Dependent Sample (EDS). Journal of Animal Ecology, vol. 71, pp. 1015-1029.
MILLER, M.P., 2005. Alleles In Space: Computer software for the joint analysis of interindividual spatial and genetic information. The Journal of Heredity, vol. 96, no. 6, pp. 722-724. http://dx.doi. org/10.1093/jhered/esi119. PMid:16251514.
MÜLLER, L., GONÇALVES, G.L., CORDEIRO-ESTRELA, P., MARINHO, J.R., ALTHOFF, S.L., TESTONI, A.F., GONZÁLEZ, E.M. and FREITAS, T.R., 2013. DNA Barcoding of Sigmodontine Rodents: Identifying Wildlife Reservoirs of Zoonoses. PLoS One, vol. 8, no. 11, pp. 1-12. http://dx.doi.org/10.1371/journal. pone.0080282. PMid:24244670.
NIELSEN, R., 2001. Statistical tests of selective neutrality in the age of genomics. Heredity, vol. 86, no. Pt 6, pp. 641-647. http:// dx.doi.org/10.1046/j.1365-2540.2001.00895.x. PMid:11595044.
PAGLIA, A.P., FONSECA, G.A.B., RYLANDS, A.B., HERRMANN, G., AGUIAR, L.M.S., CHIARELLO, A.G.C., LEITE, Y.L.R., COSTA, L.P., SICILIANO, S., KIERULFF, M.C.M., MENDES, S.L., TAVARES, V.C., MITTERMEIER, R.A. and PATTON, J.L. 2012. Lista Anotada dos Mamíferos do Brasil/Annotated Checklist of Brazilian Mammals. 2. ed. Arlington: Conservation International, 76 p. Occasional Papers in Conservation Biology, no. 6.
PINHO, A.P., CUPOLILLO, E., MANGIA, R.H., FERNANDES, O. and JANSEN, A.M., 2000. Trypanosoma cruzi in the sylvatic environment: distinct transmission cycles involving two sympatric marsupials. Transactions of the Royal Society of Tropical Medicine and Hygiene, vol. 94, no. 5, pp. 509-514. http://dx.doi.org/10.1016/
S0035-9203(00)90069-5. PMid:11132378.
QUINTELA, F.M., GONÇALVES, B.I., TRINDADE, G.E., SANTOS, M.B. and TOZETTI, A.M., 2013. Non-volant small
mammals (Didelphimorphia, Rodentia) in coastal grasslands
of southernmost Brazil. Biota Neotropica, vol. 13, no. 4, pp. 284-289. http://dx.doi.org/10.1590/S1676-06032013000400025.
REIS, N.R., PERACCHI, A.L., PEDRO, W.A. and LIMA, I.P. 2006. Mamíferos do Brasil. Londrina: Universidade Estadual de Londrina, pp. 437.
ROCHA, R.G., FERREIRA, E., LOSS, A.C., HELLER, R., FONSECA, C. and COSTA, L.P., 2015. The Araguaia River as an Important Biogeographical Divide for Didelphid Marsupials in Central Brazil. The Journal of Heredity, vol. 106, no. 5, pp. 593-607. http://dx.doi.org/10.1093/jhered/esv058. PMid:26249652.
ROZAS, J., SANCHEZ-DELBARRIO, J.C., MESSENGUER,
X., and ROZAS, R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, vol. 19, pp. 2496-2427.
SAMBROOK, K.J. and RUSSEL, D.W., 2001. Molecular Cloning: A laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press.
SANCHES, V.Q.A., GOMES, M.M.A., PASSOS, F.C., GRACIOLLI, G. and RIBAS, A.C.A., 2012. Home-range and
space use by Didelphis albiventris (Lund 1840) (Marsupialia, Didelphidae) in Mutum Island, Paraná river, Brazil. Biota Neotropica, vol. 12, no. 4, pp. 50-55. http://dx.doi.org/10.1590/ S1676-06032012000400004.
SHERLOCK, I.A., MIRANDA, J.C., SADIGURSKY, M. and GRIMALDI JÚNIOR, G., 1984. Natural infection of the opossum Didelphis albiventris (Marsupialia, Didelphidae) with
Leishmania donovani, in Brasil. Memorias do Instituto Oswaldo
SMITH, T.B., 1997. A role for ecotones in generating rainforest biodiversity. Science, vol. 276, no. 5320, pp. 1855-1857. http:// dx.doi.org/10.1126/science.276.5320.1855.
SOUSA, L.C.C., GONTIJO, C.M.F., BOTELHO, H.A. and FONSECA, C.G., 2012. Mitochondrial genetic variability of Didelphis albiventris (Didelphimorphia, Didelphidae) in Brazilian
localities. Genetics and Molecular Biology, vol. 35, no. 2, pp. 522-529. http://dx.doi.org/10.1590/S1415-47572012005000035. PMid:22888303.
TAJIMA, F. 1989. Statistical methods for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, vol. 123, no. 3, pp. 585-595.
TAMURA, K., STECHER, G., PETERSON, D., FILIPSKI, A. and KUMAR, S., 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Molecular Biology and Evolution, vol. 30, no. 12, pp. 2725-2729. http://dx.doi.org/10.1093/molbev/ mst197. PMid:24132122.
THOMPSON, J.D., HIGGINS, D.G. and GIBSON, T.J., 1994. ClustaW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research, vol. 22, no. 4, pp. 4673-4680. PMid:7984417.
WANG, Z., HUBBARD, F.J., CLUBB, F.J. and VANDEBERG, J.L., 2009. The Laboratory Opossum (Monodelphis domestica)
as a Natural Mammalian Model for Human Cancer Research. International Journal of Clinical and Experimental Pathology, vol. 2, no. 3, pp. 286-299. PMid:19079623.
Supplementary Material
Supplementary material accompanies this paper.
Table S1. List of the D. albiventris specimens included in this study, their field numbers (BC, PR, FNI, ALN, TNM, T, PNSC, URC, LLLS, and DCN), GenBank accession number (JN), demes, haplotype number (H), locality,
geographical coordinates and origin of the samples.