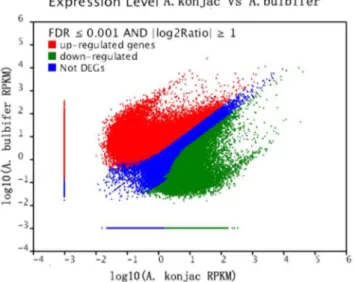
De novo transcriptome and small RNA analyses of two amorphophallus species.
Texto
Imagem




Documentos relacionados
Differentially expressed genes - Midguts from sugar and blood fed female insects were dissected and total RNA purified.The cDNA obtained by RT- PCR was separated on
The small RNAs derived from all pre-miRNA candidates (Dataset G in S1 File) were among the most abundant identified in the deep sequencing data from the H361 infection.. The 12 of
Real-time RT-PCR analyses of genes cryD , wcoA , vvdA and carRA in total RNA samples from the wild type grown for three days in DGasn medium in the dark or after 15 min, 30 min, 1 h,
Samples were collected from 1,022 asymptomatic domestic and wild birds from the Brazilian coast and the Amazon region using tracheal/cloacal swabs and tested by
Using the kit that showed better performance, two systems of Real- time RT-PCR (RT-qPCR) assays were tested and compared for analytical sensitivity to Canine Distemper Virus
Furthermore, comparison of yak and cattle ovary transcriptome data revealed that 1307 genes were significantly and differentially expressed between the two libraries, wherein 661
Differentially expressed genes after osmotic stress The four groups from the two oyster species were also separately aligned to the each reference transcriptome in order to perform
chain reaction (PCR) genotyping using species- and genotype- specific primers, nucleotide sequencing of ribosomal DNA (rDNA), proteomic analysis, and real-time PCR could be