phosphor ylation under str ess conditions
Jor ge Per eir a
Disser tation pr esented to obtain a Ph.D. degr ee in Biochemistr y
Instituto de Tecnologia Química e Biológica Univer sidade Nova de Lisboa
Supervisor Pr ofessor Claudina Rodr igues-Pousada
Oeir as, December 2008
“Com o apoio financeir o da Fundação para a ciência e Tecnologia (FCT) e do Fundo Social Eur opeu (FSE) no âmbito do Quadr o Comunitár io de Apoio,
ii
The w or k r epor ted in this thesis w as per for med in the
Genomics and Str ess Labor ator y,
Instituto de Tecnologia Química e Biológica
Univer sidade Nova de Lisboa
iii Pedr o Mor adas-Fer reira, Jorge Pereir a, Pr ofessor Claudina Rodr igues-Pousada, Pr ofessor Frédéric Devaux, Pr ofessor Cecília Arraiano and Pr ofessor Miguel Teixeira.
Members of the Jury
Professor Miguel Teixeira
(Metallopr oteins and Bioenergetics, ITQB, President of the Juri)
Pr ofessor Frédéric Devaux
(Laboratoir e de Génétique Molèculair e, CNRS, France) Professor Margarida Casal
(Depar tment of Biology, CBMA, Univer sidade do Minho, Br aga) Professor Pedro Moradas-Ferreira
(Cellular and Applied Micr obiology, IBMC, Univer sidade do Por to) Professor Cecília Arraiano
(Contr ol of Gene Expression, ITQB, Univer sidade Nova de Lisboa) Professor I sabel Sá Nogueira
(Micr obial Genetics, ITQB, Univer sidade Nova de Lisboa) Professor Claudina Rodrigues-Pousada
v Publications
Pereira, Jet al. (2008). Yap4 PKA-dependent phosphor ylation affects
pr otein stability but not its localization. Submitted to Yeast;
Nevitt T, Pereira J, Azevedo D, Guer r eir o P and Rodr igues-Pousada C
(2004). Expr ession of YAP4 in Sacchar omyces cer evisiae under
osmotic str ess. Biochem. J. 379, 367-374;
Nevitt T, Pereira J and Rodrigues-Pousada C (2004)
YAP4 gene expr ession is induced in r esponse to sever al for ms of
str ess in Sacchar omyces cer evisiae. Yeast 21, 1365-1374;
Rodr igues-Pousada C, Nevitt T, Menezes R, Azevedo D, Per eira J,
Amar al C (2004). Yeast activator pr oteins and str ess r esponse: an
over view . FEBS Letter s 567, 80-85;
vii
This w or k is dedicated to:
Idalina
ix
Antes de agr adecer a todas as pessoas que dir ecta ou indir ectamente for am impor tantes na r ealização deste tr abalho, devo confessar que esta foi a última secção da tese, e a mais difícil, a ser escr ita pois é sempr e complicado agr adecer devidamente a toda a gente sem esquecer ninguém. Por isso, deixo, desde já, as mi nhas desculpas se por omissão ou esquecimento tal acontecer .
Conheci a Pr ofessor a Claudina Rodr igues-Pousada ainda dur ante a minha Licenciatur a em Bioquímica e desde então, se dúvidas ainda havia de que ir ia enver edar pela investigação, elas desapar ecer am com o entusiasmo contagiante que a Pr ofessor a levava consigo par a as aulas. A sua deter minação e per sever ança inesgotáveis, mesmo em tempos difíceis a nível pessoal que a afligir am e continuam a afligir , influenciar am-me e inspir ar am-me, dando-me cor agem par a não desistir , mesmo quando os sacr ifícios pessoais exigidos er am demasiados. Muito mais haver ia par a dizer e agr adecer , mas tenho de ser sintético e por isso aqui fica o meu pr ofundo r econhecimento e agr adecimento à Pr ofessor a Claudina Rodr igues-Pousada por tudo quanto me ensinou e fez par a que fosse possível eu poder estar a escr ever estes agr adecimentos.
Cr onologicamente, a Tr acy Nevitt foi a pessoa com quem mais dir ectamente tr abalhei no labor atór io até à sua par tida par a os EUA. Agr adeço a sua disponibilidade e paciência par a me e integr ar no labor atór io e ensinar tudo quanto eu ainda não sabi a e er a impor tante par a tr abalhar nesta ár ea.
x
abor dagens par a fazer avançar a investigação sobr e o Yap4 quando ela par eci a estar bloqueada. Nesta r ecta final, a ajuda da Regina e da Catar ina Pimentel, quer no tr abalho labor ator ial na conclusão de algumas exper iências, quer nas discussões de resultados obtidos, for am decisivas. Muito obr igado pela vossa ajuda e disponibilidade. Conheço a Catar ina Amar al desde os tempos da Faculdade, no Por to. Foi sempr e uma pessoa r eser vada, mas igualmente disponível e voluntar iosa a ajudar -me sempr e que tal foi necessár io. Agr adeço igualmente a sua disponibilidade na r ealização de diver sas exper iências ao longo do Doutor amento.
Muitas outr as pessoas estiver am pr esentes ao longo do meu Doutor amento, or a colabor ando no meu tr abalho, or a levantando questões e suger indo caminhos a explor ar no pr osseguimento do meu tr abalho, or a inter agindo no labor atór io, cr iando um ambiente científico pr ofícuo. Agr adeço nesse sentido aos anter ior es membr os do nosso labor atór io, nomeadamente a Pr ofessor a Solange Oliveir a, a Dulce Azevedo, a Manuela Br oco, a Rute Rodr igues, a Patr ícia Machado, a Rute Félix e a Cr istina Alves, mas também aos novos elementos Fábio Silva, Ana Raposo, Joana Ropio e Cr istina Vicente. Devo igualmente agr adecer institucionalmente à FCT pela bolsa de Doutor amento atr ibuída e ao ITQB e, em par ticular , ao Labor atór io de Genómica e Str ess pelas excelentes condições de acolhimento e desenvolvimento do tr abalho de Doutor amento.
xi
nível pessoal, pr incipalmente os meus cunhados Dalila e Sousa.
E finalmente este último par ágr afo é dedicado inteir amente à pessoa que teve o papel mais impor tante nesta caminhada: a Idalina. O seu apoio e confiança e os sacr ifícios pessoais que tivemos que fazer em momentos difíceis, principalmente nos últimos tr ês anos, for am fundamentais par a que eu pudesse ter continuado e concluído o meu Doutor amento. Muito obr igado Id! Amo-te muito!
xiii
The existence of molecular mechanisms of r esponse, r epair and adaptation, many of w hich ar e gr eatly conser ved acr oss natur e, gives to the cell w ith the plasticity it r equir es to adjust to its ever -changing envir onment, a homeostatic event that is ter med the str ess r esponse. In the budding yeast Sacchar omyces cer evisiae ther e is a par ticular family of tr anscr iption factor s, the Yap family, w hich has been show n to have a r elevant r ole in yeast adaptation to sever al str ess conditions. In par ticular , Yap1 is the major r egulator of the tr anscr iptional r esponse to oxidative str ess and Yap2 and Yap8 play impor tant r oles upon cadmium and ar senic exposure, r espectively.
Another Yap member , Yap4, w as initially associated to chr omosome instability (CIN5) and r esistance to cisplatin and other antimalar ial dr ugs under over expr ession conditions. Later , both YAP4 and YAP6
w er e identified as genes that confer salt toler ance w hen over expr essed in the ena1 mutant. Under this context, w e have studied YAP4 r egulation and w e show ed that YAP4 is r esponsive to a br oad r ange of str ess conditions, including osmotic and oxidative str ess, temper atur e shift and ar senic exposur e, among other s. This is due to the existence of a pr omoter r egion that is ver y r ich in differ ent
cis-elements, including sever al Str ess Response Elements (STREs), know n to be binding sites of the tr anscr iption factor Msn2, Heat Shock Elements (HSEs) r ecognized by Hsf1, as w ell as a Yap1 Recognition Element (YRE). Upon osmotic str ess, Msn2 and Hog1 have been show n to be impor tant for tr anscr iptional r egulation of
xiv
The r esults obtained in this w or k show ed that Yap4 is a phosphopr otein highly induced upon the differ ent str ess conditions pr eviously show n to incr ease the tr anscr iption of its gene, as w ell as dur ing stationar y phase. This modification w as confir med after alkaline phosphatase tr eatment that r educes the two phosphor ylated Yap4 isofor ms w ith slow er mobility shifts to the single, unphosphor ylated and fast migr ating isofor m. The same patter n w as obtained in a null Pr otein Kinase A (PKA) mutant, show ing that Yap4 phosphor ylation is PKA-dependent. Fur ther mor e, w e sear ched for putative inter mediate kinases for this modification having obser ved that Yap4 phosphor ylation is independent of kinases that cr osstalk w ith the PKA pathw ay, namely Rim15, Sch9 and Yak1. Additionally, Hog1, the MAP kinase of the HOG pathw ay, w hich is involved in the tr anscr iption r egulation of YAP4, does not phosphor ylate Yap4. Yap4 phosphor ylation is also independent of Slt2, the PKC1 MAP kinase and of the kinases Ste20, Ptk2 and Mck1, show ed to be able to phosphor ylate Yap4 in vit r o. This pr otein has sever al putative phosphor ylation sites and S99 and S210 w er e predicted to be PKA tar gets in vivo. How ever , our data did not confirm this pr ediction, show ing that only the mutation of r esidues T192 and S196 impair s Yap4 phosphor ylation. The abolishment of phosphor ylation in both mutants is independent of the str ess conditions applied and did not affect Yap4 nuclear localization or its ability to par tially r escue the
xv
xvii
A existência de mecanismos de r esposta, r epar ação e adaptação, muitos dos quais conser vados ao longo da evolução desde a bactér ia ao Homem, dotou as células com a flexibilidade necessár ia par a se ajustar ao seu meio ambiente em constante mudança. Estes mecanismos necessár ios à manutenção da homeostasia inter na da célula constituem a r esposta ao str ess.
A levedur a Sacchar omyces cer evisiae contém uma família par ticular de factor es de tr anscr ição, designada por família Yap, que tem um papel r elevante na adaptação da levedur a a difer entes condições de str ess. Concr etamente, o Yap1 é o pr incipal r egulador da r esposta tr anscr ipcional ao str ess oxidativo e o Yap2 e Yap8 desempenham papéis impor tantes na r esposta da levedur a à exposição ao cádmio e ar sénico, r espectivamente.
Outr o membr o da família Yap, o Yap4, foi inicialmente associado à instabilidade cr omossomática (sendo designado por CIN5) e à r esistência à cisplatina e outr as dr ogas contr a a malár ia, em condições de sobr e-expr essão. Mais tar de, o YAP4 e o YAP6 for am identificados como genes que confer iam toler ância ao sal no mutante ena1, novamente quando sobr e-expr essos.
xviii
após um str ess osmótico, os factor es Msn2 e Hog1 são necessár ios par a a r egulação tr anscr ipcional do YAP4 e no caso do str ess oxidativo, esta r egulação é efectuada pelos factor es Yap1 e Msn2. Os r esultados obtidos neste tr abalho mostr am que o Yap4 é uma pr oteína fosfor ilada, sendo induzida nas difer entes condições de str ess que anter ior mente mostr amos aumentar em a expr essão do gene que a codifica, e ainda dur ante a fase estacionár ia. Esta modificação pós-tr aducional foi confir mada após tr atamento dos extr actos pr oteicos com fosfatase alcalina que conver teu as duas isofor mas fosfor iladas do Yap4 na isofor ma não fosfor ilada, com uma maior mobilidade electr ofor ética. O mesmo padrão de migr ação é obtido no mutante par a a pr oteína cinase A (PKA), mostr ando que a fosfor ilação do Yap4 é dependente desta cinase. Além disso, pesquisamos a existência de possíveis cinases r elacionadas ou não com a PKA que pudessem ser inter mediár ias entre a PKA e o Yap4. Ver ificamos que a fosfor ilação do Yap4 er a independente das cinases Rim15, Sch9 e Yak1. Mostr amos também que a cinase Hog1, a “Mytogen-Activated Pr otein” cinase (MAPK) da via de sinalização HOG de r esposta ao choque hiper osmótico, e que anter ior mente mostr amos r egular a expr essão do YAP4, não fosfor ila o Yap4. O mesmo sucedeu com a cinase Slt2, a “Mytogen-Activated Pr otein” (MAPK) cinase da via de sinalização da pr oteína cinase C envolvida na manutenção da integr idade da par ede celular . Além disso, foi mostr ado in vit r o que as cinases Ste20, Ptk2 e Mck1 fosfor ilam o Yap4, mas os nossos r esultados in vivo não confir mar am aqueles dados. A pr oteína Yap4 possui múltiplos r esíduos potencialmente fosfor iláveis, tendo sendo a S99 e a S210 pr evistas ser em fosfor ilados
xix
xxi
The w or k pr esented in this thesis is a contr ibution to the under standing of Yap4 r ole in the budding yeast Sacchar omyces cer evisiae under str ess r esponse.
In Chapter I w e r eview the state of the ar t r egar ding tr anscr iption factor s, the specific Yap pr oteins and the mechanisms of str ess r esponse and signalling in yeast.
This study star ted with the char acter ization of Yap4 expr ession upon osmotic, oxidative and other for ms of str ess pr esented as show n as show n in Chapter II.
Yap4 phosphor ylation w as investigated and the r esults ar e descr ibed in Chapter III.
The data obtained on the study of Yap4 localization ar e pr esented in Chapter IV.
xxiii 32P isotope 32 of the chemical element
ppppppppppphosphor ous AP-1 activator pr otein 1
ARE AP-1 r ecogniti on element
bp base pair s
BR basic r egion
BSA bovine ser um albumi ne
bZIP basic DNA-binding domain and leucine zi pper
cAMP adenosine-3’,5’-cyclic monophosphate
cCRD C-ter minal cysteine r ich domai n
Ci Cur ie
CIP calf intesti nal al kaline phosphatase
DABCO 1,4-diazabicyclo[2.2.2]octane
DAPI 4’,6’-diamidino-2-phenylindole, dihydr ochlor ide
dATP 2'-deoxyadenosi ne 5'-tr iphosphate
dCTP 2'-deoxycytidine 5'-tr iphosphate
dGTP 2'-deoxyguanosine 5'-tr iphosphate
DNA deoxyr ibonucleic acid
dNTP deoxy-any base-5'-tr iphosphate
DTT 1,4-dithiothr eitol
dTTP 3'-deoxythymidine 5'-tr iphosphate
EDTA ethylenediami netetr aacetic acid
ESR envir onment str ess r esponse
GFP gr een fluor escent pr otei n
GSR gener al str ess r esponse
GTF gener al tr anscr iption factor
HOG high osmolar ity glycer ol
HSE heat shock element
HSF heat shock factor
HSP heat shock pr otei n
KAN kanamycin
kb kilo base pair s
kDa kiloDalt on
LB Lur ia-Ber tani
LZ leucine zi pper
MAP mitogen activator pr otein
MAPK MAP ki nase
MOPS 3-(N-mor pholino)pr opanesulfonic acid
mRNA messenger r ibonucleic acid
n-CRD N-ter minal cysteine r ich domain
NES nuclear expor t si gnal
NLS nuclear localizati on signal
nm nanometer s
nt nucleotides
OD600nm optical density at 600 nm ºC degr ee Cel sius Oligo oligonucleotide
ORF open r eadi ng fr ame
PBS phosphate buffer ed saline
PCR polymer ase chain r eaction
PEG polyethylenoglycol
pH -log10[H+]
PKA pr otein kinase A
PSA ammonium per sulfate
RNA r ibonucleic acid
ROS r eactive oxygen species
r pm r otations per minute
xxiv
S. cer evisiae Sacchar omyces cer evisiae
S. pombe Schizosacchar omyces pombe
SDS sodium dodecyl sulphate
STRE str ess r esponsive element
TF tr anscr ipti on fact or
YRE Yap r esponse element
Amino acids
A Ala alanine
B Asx aspar agine or aspar tic
aaaaaaaaaaaaaaaaaaaaacid C Cys cysteine
D Asp aspar tic acid
E Glu glutamic aci d
F Phe phenylalanine
G Gly glycine
H His histidine
I Ile isoleuci ne
K Lys lysine
L Leu leucine
M Met methioni ne
N Asn aspar agine
P Pr o pr oline
Q Gln glutamine
R Ar g ar ginine
S Ser ser ine
T Thr thr eonine
V Val valine
W Tr p tr yptophan
Y Tyr tyr osine
Z Glx glutamine or
. glutamat e X X any aa r esi due li sted
Nucleotide bases
A adenine
C cytosine
G guanine
T thymine
N any base li sted above
1 I I ntroduction
I .1 Transcription factors
I.1.1 The differ ent families of Transcription Factor s………... 7 I.1.2 AP-1 tr anscr iption factor s and the Yap family………... 13 I .2 Role of the YAP factors in the stress response
I.2.1 Yap1 and Yap2………... 18 I.2.2 Yap8………... 23 I.2.3 Yap3, Yap5 and Yap7………... 24 I.2.4 Yap4 and Yap6………... 26 I .3 The Environment Stress Response... 29 I.3.1 The Gener al Str ess Response...…………... 32 I.3.2 The heat shock r esponse... 36 I.3.3 The PKA pathway………... 43 I.3.4 The HOG pathw ay………... 47 I I YAP4 regulation under different stress conditions.…….……... 57 I I I Role of Yap4 phosphorylation………... 79 I V Determinants of Yap4 localization………...… 105 V Conclusions and perspectives………... 125 Annex
A. Alignment of YAP family………... 133 B. Mater ial and Methods………... 141 C. List of str ains...…...………... 163 D. List of oligomer s...………... 165 E. List of plasmids………... 167 F. Publications………... 169 References... .
3 Fig. 1.1: DNA-binding and dimer ization domains of the major
eukar yotic TF families………...………. 12 Fig. 1.2: Scheme of a leucine zipper (LZ) par allel tw o-str anded
coiled coil and side view………...……….. 13 Fig. 1.3: Yeast tr anscr iption factor Gcn4-bZIP binding to DNA... 14 Fig. 1.4: Alignment and compar ison of GCN4 basic r egion and
Leucine Zipper w ith the YAP family member s…………... 17 Fig. 1.5: Over view of Yap1 r egulation thr ough nuclear expor t... 22 Fig. 1.6: Over view of YAP4 and YAP6 cis-elements………... 29 Fig. 1.7: Over view of yeast ESR major player s and pathw ays…... 31 Fig. 1.8: Over view of Msn2 activity via its phosphor ylation
dependent sub-cellular localization…... 36 Fig. 1.9: Gener ic str uctur e of Hsf1 in yeast and mammalian cells... 37 Fig. 1.10: Over view of PKA pathway cr osstalk with GSR thr ough Msn2 46 Fig. 1.11: Over view of the S. cer evisiae HOG pathw ay... 54 Fig. 2.1: Over view of YAP4 r egulation under differ ent str ess
conditions... 62 Fig. 2.2: YAP4 is r esponsive to osmotic str ess... 63 Fig. 2.3: Yap4 is induced and phosphor ylated upon osmotic str ess... 65 Fig. 2.4: Hog1 affects Yap4 levels but not its phosphor ylation... 66 Fig. 2.5: Expr ession analysis of DCS2, GPP2 and GCY1 genes in w t
and yap4 mutant str ains... 67 Fig. 2.6: YAP4 deletion affects the expr ession of Dcs2... 69 Fig. 2.7: Inter nal glycer ol accumulation is unaffected by the lack of
Yap4... 70 Fig. 2.8: YAP4 induction under oxidative str ess is dependent on
Yap1 and Msn2... 71 Fig. 2.9: Yap4 is tr ansiently induced and phosphor ylated under
oxidative str ess... 72 Fig. 2.10: Yap4 levels ar e sever ely affected in a bcy1 str ain... 73 Fig. 2.11: S196A mutation pr events Yap4 phosphor ylation and S89
4
Fig. 3.1: Kinetics of Yap4 phosphor ylation………... 85 Fig. 3.2: Schematic r epr esentation of pr edicted Yap4 str uctur al
domains... 86 Fig. 3.3: Yap4 phosphor ylation is dependent on PKA... 89 Fig. 3.4: S196A mutation abolishes Yap4 phosphor ylation... 91 Fig. 3.5: T192A, S196A and T192AS196A mutations abolish Yap4
phosphor ylation under differ ent str ess conditions... 92 Fig. 3.6: Non-phosphor ylated Yap4 localizes in the nucleus... 95 Fig. 3.7: Absence of phosphor ylation does not compr omise Yap4
ability to r escue the hog1 osmosensitive phenotype... 96 Fig. 3.8: Absence of Yap4 phosphor ylation does not seem to affect
the expr ession of Dcs2... 97 Fig. 3.9: Absence of Yap4 phosphor ylation incr eases the HXT5
expr ession... 97 Fig. 3.10: Yap4 stability is par tially dependent on phosphor ylation... 99 Fig. 4.1: Yap4 impor t into the nucleus is not mediated by the
impor tin Pse1... 111 Fig. 4.2: Schematic r epr esentation of pr edicted Yap4 str uctur al
domains... 113 Fig. 4.3: Mutation of Yap4 basic r esidues of its pr edicted NLSs does
not pr event its nuclear localization... 115 Fig. 4.4: Yap4 phosphor ylation is impair ed in the
R193AK194AK242AR243AYap4 and K31AK32AR193AK194AYap4 mutants... 116
Fig. 4.5: Mutation of Yap4 NLS key r esidues compr omises its ability to r escue the hog1 osmosensitive phenotype... 116 Fig. 4.6: Yap4 deletion analysis... 118 Fig. 5: Over view of the differ ent mechanisms that r egulate Yap4... 126 Tables
5
Intr oduction
I . I ntroduction
I .1 Transcription factors
I.1.1 Character ization of the differ ent families of Tr anscr iption Factor s... 7 I.1.2 AP-1 tr anscr iption factor s and the Yap family………... 13 I .2 Role of the YAP factors in the stress response
I.2.1 Yap1 and Yap2………... 18 I.2.2 Yap8………... 23 I.2.3 Yap3, Yap5 and Yap7………... 25 I.2.4 Yap4 and Yap6………... 26 I .3 The Environment Stress Response
7
I. Intr oduction
I.1 Tr anscr iption factor s
I.1.1 Characterization of the different families of Transcription Factors
Gene tr anscr iption and its r egulation in eukar yot ic cells is a highly coor dinated pr ocess, involving hundr eds of pr oteins or ganized in differ ent complexes. DNA sequence-specific pr oteins, know n as tr anscr iption factor s (TFs), ar e able to decipher the tr anscriptional r egulator y code by binding par ticular DNA sequences (binding sites know n as cis-r egulator y elements) in the pr omoter r egion of the tar gets genes. This differ ential occupancy of the pr omoter r egions by the differ ent TFs w ill favour the assembly and disassembly of the tr anscr iption machiner y (Levine and Tjian, 2003) and consequently the expr ession levels of the tar gets genes.
8
TF can be divided accor ding to their function in thr ee major gr oups: the Gener al Tr anscr iption Factor s (GTFs: TFIIA, -B, -D, -E, -F, and –H), the induced sequence-specific TFs and the chr omatin r emodelling and modification associated TFs, also designated tr anscr iption co-factor s (Fazzio et al., 2001; Er kina et al., 2008; Steinfeld et al. 2007). The GTFs ar e basal TFs, acting at the pr imar y level of tr anscr iption and they ar e necessar y for the pr omoter r ecognition and cor r ect r ecr uitment of RNA polymer ase II and other associated r egulator s. The sequence-specific DNA binding TFs ar e r egulator y TFs that, unlike GTFs, bind to sequences fur ther aw ay fr om the initiation site and ser ve to modulate the tr anscr iption of the tar gets genes. TFs expr ession and activation is also modulated as consequence of differ ent cell challenges. The chr omatin r emodelling and modification associated TFs ar e pr esent in complexes that assist the tr anscr iptional appar atus to navigate thr ough chr omatin (Levine et al., 2003).
Fr om a str uctur al point of view , TFs ar e modular pr oteins, possessing a DNA-binding domain (DBD) that w ill bind dir ectly to the DNA and a tr ans-activating domain w hich contain binding sites for other pr oteins, such as tr anscr iption co-r egulator s, that w ill allow the r ecr uitment of the basal tr anscr iption machiner y (Kadonaga, 2004). In addition, TFs can also possess one or mor e activation or r epr ession domains w ith a signal sensing motif r esponsible for the sensing of exter nal signals thr ough the binding of a ligand. In many cases, how ever , the DBD and the signal sensing domains belong to differ ent pr oteins that associate w ithin the tr anscr iption complex to r egulate gene expr ession (Amoutzias et al., 2008).
9
homo or heter odimer ize, give r ise to many new TF dimer s w ith distinct DNA binding pr operties. Consequently many novel genetic r egulator y netw or ks that w ill allow a fine-tuning of gene expr ession ar e gener ated. Regar ding the heter odimer ization, the concentr ation of each monomer in the cell, the possibility of post -tr anslational modifications, such as phosphor ylation, and differ ent binding affinities for other monomer s w ill deter mine w hich dimer (heter o or homodimer ) w ill be for med and, consequently, w hich signalling pr ocess w ill pr evail over the other s. This is the case of the Myc-Max and Mad-Max heter odimer ization system that defines w hether a lar ge number of tar geted genes w ill be expr essed or silenced. The TFs Jun and ATF2 ar e another example as they pr esent differ ent binding activities fr om the Jun or ATF2 homodimer s (Amoutzias et al., 2008 and r efer ences w ithin). The case of Jun and Fos is also par adigmatic as each of them for ms homodimer s but only the heter odimer is functional (O’Shea et al., 1992).
Consider ing the TFs that ar e able to dimer ize, a ver y heter ogenic gr oup w ith sever al motifs that char acter ize the DBD and define differ ent families w as descr ibed (r eview ed in Amoutzias et al., 2008 and r efer ences ther ein). The gener al str uctur e of the most r elevant families of TF that exer t their function as dimer s is r epr esented in Fig. 1.1.
The bHLH (basic-r egion helix–loop–helix) family of TFs is the lar gest family of dimer izing TFs in humans. Many bHLH TFs ar e impor tant r egulator s of development, differ entiation and cell cycle. These TFs ar e char acter ized by an alpha-helical basic r egion (BR) that binds E-box DNA elements (CANNTG) as a dimer . The helix–loop–helix (HLH) for ms a four -helix bundle and is r esponsible for the dimer ization (Amoutzias
10
HLH. This second dimer ization domain confer s higher specificity in the dimer ization pr ocess.
The bZIP (basic r egion leucine zipper ) is the second-lar gest family of dimer izing TFs in humans, many of them being the w ell-studied oncogenes. They can be gener ically consider ed as envir onmental biosensor s and contr oller s of development, r egulating development, metabolism, cir cadian r hythm, lear ning, memor y, and r esponse to str ess and r adiation (Amoutzias et al., 2008 and r efer ences w ithin). bZIPs TFs str uctur e is ver y similar to bHLH TFs, being the alpha-helical basic r egion r esponsible for the binding to the DNA. Again, dimer ization occur s via the C-ter minal coiled-coi l LZ (Vinson et al., 2006). Fig. 1.2 highlights the str uctur e of a bZIP dimer , focusing the r esidues that ar e impor tant for dimer ization.
The HD-ZIP (Homeodomain leucine zipper ) is a thr ee alpha helices helix–tur n–helix DNA-binding domain specific to plants. In this family of TFs, DNA binding is mediated by the homeodomain and the thir d helix confer s DNA-binding specificity. Again, the LZ domain dir ects dimer ization (Tr on et al., 2004).
In the MADS box family of TFs, the MADS domain dir ects DNA binding and the domains that mediate dimer ization ar e located C-ter minal to the MADS domain. This family is highly r epr esented in plants, being associated to the contr ol of or gan development (de Folter et al., 2005).
11
immunoglobulin r egion is mainly involved in dimer ization (Chen and Ghosh, 1999). NFAT (Nuclear factor of activated T cells) family of TFs is str uctur al ver y similar to the NF-kB family.
NR (Nuclear r eceptor ) family of TFs ar e of incr easing inter est since they ar e specific for humans and ar e being studied by the phar maceutical industr y as dr ug tar gets for many human diseases. In the NRs, zinc-finger (Zf) domains mediate DNA binding, w hereas dimer ization is mediated by both the Zf domain and a ligand-binding domain (Ger main
et al., 2006).
12
Figure 1.1: DNA-binding and dimerization domains of the major eukaryotic TF
families.
For simplicit y, only the DNA-binding domains (red boxes), dimerization domains (orange boxes) and addit ional dimerizat ion domains t hat confer specificit y (blue boxes) are indicated. The elements indicat ed in t he green boxes are domains t hat have both DNA-binding and dimerizat ion act ivit y. bZIP - basic region leucine zipper TF family, bHLH - basic region helix-loop-helix TF family; BR – basic region; HLH – helix-loop-helix domain; HD – homeodomain, HD-ZIP – homeodomain leucine zipper TF family; I - specific domain of a subfamily of M ADS-Box prot eins; IG - immunoglobulin; K - specific domain of a subfamily of M ADS-Box prot eins; LBD - ligand-binding domain; LZ – leucine zipper; M ADS - named aft er t he four originally identified members: MCM 1, AGAM OUS,
13 I.1.2 AP-1 transcription factors and the Yap family
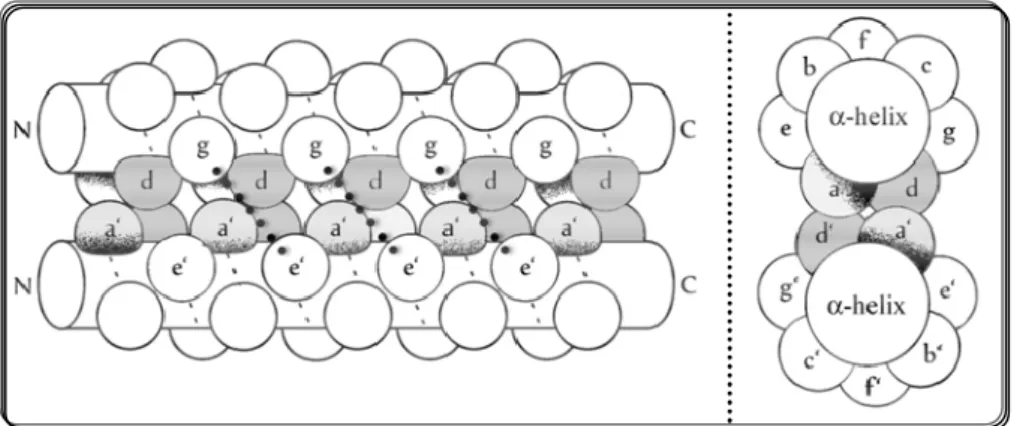
bZIP-TFs ar e par ticular TFs as their main str uctur e contains a DBD bZIP that w as deduced fr om its primar y amino acid sequence and compr ises a basic r egion (BR) and a leucine zipper (LZ). These TF dimer ize thr ough their leucine r epeats and for m a coiled coil. This consists of tw o or mor e αhelices that w ind ar ound one another w ith a slight left -handed super helical tw ist. A char acter istic heptad r epeat (abcdefg)n defines the placement of r esidues in each helix r elative to the inter action inter face (r eview ed in Fong et al., 2004) . Usually, r esidues a and d ar e hydr ophobic amino acids, w hile the exposed r esidues g and e ar e char ged and polar amino acids (Fig. 2). Adjacent to the LZ dimer ization domain is the highly conser ved BR that contacts specific hexamer s of DNA bases. bZIP w er e named accor ding to the fact that the cor e d positions ar e usually leucine (or leucine-like) r esidues (as can be seen in Fig. 1.3).
14
Whilst this appar ent simplification of bZIP sequence that allow s their easy identification and pr ediction of binding affinities, str uctur e and ener getics of their hydr ophobic inter faces thr ough molecular modelling, bZIPs TFs exhibit a high degr ee of par tner ing selectivity. This could explain w hy they ar e able to par ticipate in the r egulation of differ ent pathw ays (Fong et al., 2004). This selectivity is r eflected in the DNA binding sites that bZIPS ar e able to r ecognize, usually shor t palindr omic or pseudo-palindr omic tar get sequences. The metazoan bZIPs can r ecognize 6 differ ent consensus DNA-binding sites (the TPA r esponsive element (TRE), AMP r esponsive element (CRE), CAAT box, AF r ecognition element (MARE), CRE-like, and PAR binding sites), w hile yeast bZIPs ar e able to bind only thr ee consensus DNA-binding sites (the TRE-, CRE-, and YAP-binding sites) (Deppmann et al., 2006).
Figure 1.3: Yeast transcription factor Gcn4-bZIP binding to DNA.
15
Ther e is a par ticular gr oup of bZIP TFs, know n as AP-1 TFs, that ar e able to r ecognize the ARE (AP-1 Responsive Element, also know n as TPA-r esponsive element- TRE) TGACTCA nucleotide sequence, later designated as AP-1 site. AP-1 TFs ar e involved in sever al impor tant cellular pr ocesses in mammalians, like cell pr oli fer ation, apoptosis, development and str ess r esponse, among other s (Toone and Jones, 1999). The pr ototype AP-1 TF in the yeast Sacchar omyces cer evisiae is Gcn4 (Fig. 3). This TF pr imar ily r egulates yeast r esponse to amino acid star vation (Natar ajan et al., 2001; Hinnebusch, 2005) and is ver y similar to mammali an AP-1 factor s Jun and Fos oncopr oteins. The thr ee TFs shar e the same DNA-binding specificity (Str uhl, 1987) and they ar e able to per for m the same functions in yeast and mammalian cells (Str uhl, 1988).
Yap1, the fir st member of the family of Yaps to be descr ibed in yeast, w as initially identified by its ability to bind and activate the SV-40 AP-1 r ecognition element (ARE: TGACTAA) (Rodr igues-Pousada et al., 2004). Yap2 w as isolated in a scr eening confer r ing resist ance to 1,10-phenanthr oline in tr ansfor med cells over expr essing a yeast libr ar y and also binds ARE cis-acting element. The sequencing of the YAP1 and
16
Yap member s also contain a glutamine at position 234 and an alanine at position 241 and these tw o modifications ar e not found in any other bZIP pr oteins (r eview ed in Rodr igues-Pousada et al., 2004).
In
tr
o
d
u
c
tio
n
1
7
18
I.2 Role of the Yap factor s in the str ess r esponse.
I.2.1 Yap1 and Yap2
Yap1 is the major r egulator of the oxidative str ess r esponse. Initial studies show ed that yap1 mutants w er e hyper sensitive to the oxidants H2O2 and t-BOOH, and to chemicals that gener ate super oxide anions,
such as menadione, plumbagine, methylvi ologen, cadmium,
19
20
intr amolecular disulfide bond is for med betw een the n-CRD C310 and the c-CRD C629 upon exposur e to H2O2. Mor e r ecently, Okazaki et al., (2007) show ed that, in fact, ther e is a multistep disulfide bond for mation in Yap1 involving the cysteines for m both nCRD and cCRD domains upon exposur e to oxidative str ess imposed by H2O2.
Under oxidative str ess induced by diamide and other thiol-r eactive agents, Yap1 c-CRD is sufficient to mediate str ess r esponse, w hich suggests that Yap1 uses differ ent r edox center s to deal w ith differ ent str ess conditions. The mechanism of r esponse to oxidative str ess induced by diamide and other thiol-r eactive agents does not r equir ed Ybp1 or the per oxidase Or p1, possibly involving a dir ect binding of these oxidative str ess agents to the c-CRD cysteines C598, C620 and C629. (For r evi ew see Rodr igues-Pousada et al., 2005). In fact, this mechanism w as alr eady show n in yeast exposed to the oxidative str ess induced by N-ethylmaleimide and menadione (Azevedo et al., 2003). These r esults suggested the pr esence of at least tw o differ ent r edox center s in Yap1: one activated upon oxidative str ess imposed by H2O2 thr ough intr amolecular disulfide bonds (C303-C598 and C310-C629) and another upon exposur e to thiol-r eactive agents that bind to the cCRD cysteines (C598, C620 and C629).
21
Cell exposur e to cadmium str ess induces the expr ession of YAP2
(Fer nandes et al., 1997) and the encoded pr otein is tr anslocated to the nucleus thr ough a Cr m1-dependent mechanism, activating the tr anscr iption of its tar get gene FRM2 (Azevedo et al., 2007). This gene encodes a pr otein homologous to nitr or eductase, but is not clear w hat it is its r ole in metal str ess r esponse. Yap2 shar es a high amino acid sequence homology w ith Yap1, namely in its bZIP and C-ter minal CRD, conser ving all thr ee cysteine r esidues of Yap1 c-CRD and the hydr ophobic r esidues that compose the NES. Taking into account this homology, Azevedo et al. (2007) per for med the c-CRD sw apping betw een Yap1 and Yap2, show ing that the Yap2 containing Yap1 c-CRD w as induced by cadmium but not by H2O2. Fur ther mor e it suppr esses
22
Stress conditions
Nucleus Cytoplasm
Figure 1.5: Overview of Yap1 regulation.
Under normal grow th conditions, Yap1 circulat es bet ween nucleus and cytoplasm t hrough, respect ively, t he int eract ion bet ween its NLS and t he import in Pse1 and it s NES and exportin Crm1. How ever, exposure to oxidant conditions (H2O2) modifies Yap1 st ruct ure t hrough int ramolecular disulfide bond format ions between it s redox centers t hat mask the NES and blocks Yap1 interaction w it h Crm1, t hus accumulat ing Yap1 in t he nucleus. Additionally, Yap1 can be ret ained in t he nucleus by t hiol-react ive agents t hat form adducts with the C-t erminal cysteines and mask t he NES, again preventing it s int eract ion w it h Crm1.
23 I.2.2 Yap8
YAP8 is the most diver gent member of the Yap family. It is the main r egulator of the ar senic detoxification pathw ay, for ming a cluster (known as ar senic compounds-r esistance - ACR) w ith the genes that encode the ar senate-r eductase Acr 2 and the plasma membr ane ar senite efflux pr otein Acr 3 (Wysocki et al., 1997). In r esponse to ar senate exposur e, Yap8 is activated and induces the expression of ACR2 and
ACR3 (r eview ed in Rodr igues-Pousada et al., 2004). Ar senic compounds can be unspecifically inter nalized by yeast cells in t w o for ms: ar senate (AsV), thr ough phosphate tr ansporter s as Pho87, and ar senite (AsIII), thr ough the aquaglycepor in Fps1 or the hexose per meases Hxt1, Hxt3, Hxt4, Hxt5, Hxt7 and Hxt9. The mechanism of detoxification of ar senate involves the r eduction of ar senate to ar senite by the ar senate-r eductase Acr 2 follow ed by the extr usion of the ar senite compounds out of the cell mediated by the Acr 3 plasma membr ane ar senite efflux pr otein. A secondar y pathw ay of ar senic detoxification involves GSH conjugation of ar senite [As(GS)3] follow ed by its accumulation in the vacuole mediated by the pump Ycf1 ABC ATPase (r eviewed in Bhattachar jee and Rosen, 2007).
Yap8 is constitutively expr essed, being its activity mainly r egulated at the level of its pr otein subcellular localization. Upon exposur e to ar senic, Yap8 r apidly accumulates in the nucleus, and this tr anslocation is mediated by the expor tin Cr m1, as it happens w ith the r elated Yap1 (Menezes et al., 2004). In fact, the r egulation of Yap8 nuclear accumulation is trigger ed by the loss of inter action w ith Cr m1. Menezes
24
It is not known yet if this modification involves dir ect binding of the ar senite to the r efer r ed cysteines as it happens w ith the binding of thiol-r eactive agents to the c-CRD cysteines of Yap1 (Azevedo et al., 2003). It w as thought that Yap1 and Yap8 could exer t over lapping functions in the ar senic str ess r esponse as YCF1 induction is dependent on Yap1, as w ell as the ACR genes that ar e also par tially r egulated by Yap1 (Menezes et al., 2004). Mor eover , yap1 yap8 double mutant is mor e sensitive to ar senic conditions than any of the single mutant (Menezes et al., 2004). How ever , in a r ecent w or k, Menezes et al. (2008) clar ified Yap1 contr ibution to ar senic str ess r esponses, show ing that Yap1 is necessar y for the pr evention of oxidative damage in cells exposed to ar senic compounds, namely thr ough the r emoval of ROS gener ated.
I.2.3 Yap3, Yap5 and Yap7
Yap3, Yap5 and Yap7 ar e the least char acter ized tr anscr iption factor s of the Yap family. Yap3 show s vir tually no r esponse at the level of genomic micr oar r ay analyses to the multiple for ms of envir onmental insults and cellular str ess studied so far . How ever , Yap3 has a str ong tr ansactivation potential even gr eater than the one of Yap1 and binds the consensus YRE, TTAC/ GTAA (Fer nandes et al., 1997). Mor eover
yap3 mutant str ain exhibits a high content of glycogen, w hich suggest s that Yap3 could be r elated to the metabolism contr ol (Wilson et al., 2002).
Yap5 has been r ecently show n to be str ongly induced under nitr ogen depletion and dur ing stationar y phase, but also amino acid star vation
25
immunopr ecipitation (ChIP) and genomic micr oar r ay hybr idizations, Hor ak et al. (2002) found that YAP5 is a tar get of t he SBF (Sw i4–Swi6 cell cycle box binding) tr anscr iption factor and dow nstr eam SBF, Yap5 has sever al putative gene targets (237 in total) involved in differ ent mechanisms, such as DNA r eplication and monitor ing of the fidelity of r eplication, DNA damage r esponse (RAD4) and chromatin r emodelling (SIR4), ener gy gener ation and amino acid metabolism (12 tar gets genes), as w ell as gamma-glutamyl kinase PRO1 and the cyclin-dependent kinase Pho85. Mor eover , Yap5 binds many pr omoter s adjacent to genes w ith peak tr anscr ipt levels in G1 phase and r elated to this, tar gets CDC47, that encodes one of a six pr otein complex (MCM) r equir ed for cell cycle pr ogr ession and DNA r eplication initiation and elongation (Tye, 1999). Mollapour et al. (2004) also show ed that yap5
mutant is mor e r esistant to sor bate exposur e.
Mor e impor tantly, Yap5 w as show n to be the tr anscr iption factor that r egulates ir on metabolism, namely its vacuolar stor age (Li et al., 2008). Yap5 w as show n to r egulate the tr anscr iption of CCC1 (the major facilitator of ir on impor t into the vacuole, w hich is the most impor tant site of ir on stor age in fungi and plants). Mor eover , this ir on stor age is stimulated by ir on exposur e thr ough a Yap consensus site in the CCC1
pr omoter . Yap5 is constitutively localized in the nucleus and occupies the CCC1 pr omoter independent of the ir on concentr ation, being the
26
ther efor e its putative disulfide bonds ar e not involved in the r egulation of its expor t as in the case of Yap1, Yap2 and Yap8.
YAP7 is up-r egulated under conditions of nitr ogen depletion and stationar y phase (Gasch et al., 2000) but its function is far fr om being under stood. Our r ecent tr anscr iption pr ofiles using the double mutant
yap1 yap8 and the r espective single mutants against the w ild type, all tr eated w ith ar senate, r eveals that Yap7 seems t o be dependent on Yap1.
I.2.4 Yap4 and Yap6
Yap4 and Yap6 ar e the closest r elated Yap family member s. Yap4 is a 33kDa pr otein shar ing almost 33% identity (Rodr igues-Pousada et al., 2004) w ith Yap6 and, unlike other Yap member s, as Yap1, Yap2 and Yap8, they ar e constitutively located in the nucleus (Fur uchi et al., 2001). In addition, both Yap4 and Yap6 pr oteins w er e associated to r esistance to sever al dr ugs, including antimalar ial dr ugs, chlor oquine,
quinine and mefloquine (Delling et al., 1998), and the
27
r ecover ed by YAP4 over expr ession, show ing that Yap4 plays a r ole in the yeast r esponse to osmotic str ess.
In fact, under osmotic str ess, YAP4 Msn2-mediated induction occur s in a Hog1-dependent manner thr ough at least tw o STREs pr esent in its pr omoter r egion (Nevitt et al., 2004a). YAP4 is also r esponsive to oxidative str ess and in this case its r egulation is dependent on Yap1 and Msn2 via its YRE and most pr oximal STRE, r espectively (Nevitt et al., 2004b). In both cases, the pr otein is tr ansiently expr essed and phosphor ylated.
Genome w ide appr oaches r evealed that besides osmotic and oxidative str ess, YAP4 and YAP6 ar e induced under other str ess conditions, including heat and stationar y phase (Gasch et al., 2000; Rep et al., 2000; Posas et al., 2000). This r esult cor r elates w ell w ith the fact that both genes possess pr omoter r egions r ich in differ ent cis-elements (Fig. 1.6) , suggesting that both YAP4 and YAP6 r espond to multiple signalling pathw ays.
Differ ent computational inter actome data pr edicts the inter action betw een Yap4 and Yap6, such as the one obtained by Lee et al. (2002). How ever , the possibility that this inter action occur s via heter odimer ization r emains to be demonstr ated. Data fr om Deppmann
28
Figure 1.6: Overview of YAP4 and YAP6 cis-elements.
29
I.3 The Envir onment Str ess Response
Yeast cells ar e continuously exposed to envir onmental cues that affect its viability, such as dr astic changes in osmolar ity, temper atur e, pH, nutr ients and oxygen availability. In or der to sur vive, yeasts develop sever al mechanisms to cope with these str ess conditions. Altogether , these mechanisms can be consider ed as par t of the yeast Envir onmental Str ess Response (ESR). Msn2/ 4 and Hsf1 (Gener al Str ess Response), PKA (cAMP pathw ay), Hog1 (HOG pathw ay) and Pkc1 (pr otein kinase C-mediated MAP kinase pathw ay) ar e the key player s of some of the r egulator y pathw ays in Sacchar omyces cer evisiae as r epr esented in Fig. 1.7.
The basic mechanisms to r espond to str ess involves sensor pr oteins, usually tr ansmembr anar pr oteins associated w ith the yeast cell w all, that ar e able to sense the str ess condition and initiate a cascade of activation of dow nstr eam effector s enabling cells to cope w ith the str ess effects. These mechanisms ar e specific for the natur e of the str ess cells sense and many of them ar e conser ved along eukar yotes, although some over lapping betw een differ ent signalling pathw ays may occur . The main r eason for this over lap is the fact that differ ent str ess conditions can pr oduce the same effects in the cell (for instance, UV exposur e and heat can both pr oduce oxidative stress and w ill involve some common player s in the yeast r esponse).
30
2002). In contr ast, less than half of the induced genes have been yet functionally char acter ized and ar e associated w ith sever al cellular pr ocesses, including car bohydr ate metabolism, defence fr om oxidative str ess, cell w all modification, pr otein folding and degr adation, cytoskeletal r eor ganization, DNA r epair , fatty acid metabolism, metabolite tr anspor t, vacuolar and mitochondr ial functions, autophagy and intr acellular signalling (r eview in Gash, 2002).
31
activator , inducing sever al differ ent genes to pr otect cell fr om osmotic damage (Pr oft and Str uhl, 2002). Among these genes is MSN2/ 4 encoding one of the key player s of the GSR, w hich in tur n pr omotes the pr otection to sever al other s str ess conditions, such as heat shock, oxidative str ess or car bon sour ce star vation (Pr oft et al. 2005). Ber r y et al. (2008) w er e able to show that Msn2 and Msn4 have much mor e specific r oles in ESR than pr eviously thought, being MSN4 induced in r esponse to a var iety of str esses, w hile MSN2 is prefer entially induced upon osmotic challenge. Mor eover , both factor s display differ ent phosphor ylation pr ofiles in r esponse to differ ent conditions (Gar r eau et al., 2000), although it is not know n w hat ar e the consequences of such differ ences.
Figure 1.7: Overview of the Environment Stress Response (ESR) with the major
players and pathways in which they are involved.
32
I.3.1 The Gener al Str ess Response
The specific cr oss-pr otection elicited by Msn2 and Msn4 in the ESR can be par ticular ly consider ed as the Gener al Str ess Response (GSR) and involves a subset of ar ound 200 genes w hose expr ession is specifically alter ed in under the Contr ol of Msn2/ 4 (Gasch et al. 2000; Causton et al., 2001). The r egulation of these genes by Msn2/ 4 is achieved thr ough its binding to the cis-element CCCT, designated as Str ess Responsive Element (STRE) (Mar tinez-Pastor et al., 1996). How ever , ther e ar e some genes involved in the GSR that contain this cis-element, as the sodium pump ENA1, but that ar e unr esponsive to Msn2/ 4 (Alepuz et al.,
1997), suggesting the existence of an alter native pathw ay of r egulation for these cases.
The Msn2/ 4 factor s ar e r egulated at sever al levels of post -tr anslational contr ol, since its mRNA and tur nover r emain stable thr ough shor t per iods of str ess (De Wever et al., 2005). These for ms of r egulation include Mns2/ 4 subcellular localization and highly cor r elated state of phosphor ylation, stability and degr adation and eventually inter actions w ith r egulator y par tner s. Concer ning the r egulation of their localization, both pr oteins contain NES and NLS and ar e continuously shuttling betw een nucleus and cytoplasm thr ough an efficient and oscillator y nuclear tr anspor t system that is able to discr iminate the intensity of the str ess insults (Jacquet et al., 2003) and is modulated by car bon sour ce avai lability. In unstr essed cells, Msn2/ 4 localizes pr edominantly in the cytoplasm. How ever , upon exposur e to str ess, Msn2/ 4 r apidly r elocalize into the nucleus, pr omoti ng the tr anscription of its tar gets genes (Gor ner et al., 2002 and Jacquet et al., 2003). This
nuclear localization is pr evented by a PKA-dependent
33
the PKA pathw ay over the GSR. As such, under nor mal gr ow ing cells, PKA activity is high and Msn2 is maintained in the cytoplasm (Gor ner et al., 1998 and 2002; Gar mendia-Tor r es et al., 2007). How ever , under str ess conditions, PKA activity decr eases and consequently the level of phosphor ylation of Msn2/ 4 diminished, r etaining the tr anscr iption factor in the nucleus. Msn2 possesses four PKA-consensus sequences w ithin t he NLS and one within t he NES, including the ser ine r esidues S582, S620, S625, S633 and S288 r espectively, that w er e show n to be phosphor ylated under differ ent str ess conditions (Gor ner et al., 2002 and De Wever et al., 2005). These r esidues ar e inter calated by basic ar ginines in both signals and most pr obably ser i ne phosphor ylation affects the r ecognition of Msn2 NLS by the kar yopher ins involved in its tr anslocation, ther efor e r egulating Msn2 distribution and activity. This model is suppor ted by differ ent obser vations. Firstly, inactivation of PKA leads to the per manent localization of Msn2 w ithin the nucleus (Gor ner et al., 2002; Jacquet et al., 2003). Secondly, Msn2 nuclear impor t is favour ed by the dephosphor ylation of the ser ine r esidues in the NLS (De Wever et al., 2005). Finally, r eplacement of Msn2 NES (w hich contains the ser ine r esidue S288) by the pr otein kinase inhibitor (PKI) NES compr omises Msn2 expor t fr om the nucleus (Gar mendia-Tor r es et al., 2007). This also suggest s that, unlike impor tins that have higher affinity for the nonphosphor ylated for m of the Msn2, the expor tin w ould r ather pr efer entially inter act w ith the NES-phosphor ylated for m. Ther efor e one for m of ter mination of Msn2 activity w ould be its nuclear phosphor ylation and consequent expor t to the cytoplasm, w her e it is hyperphosphor ylated in a PKA-dependent manner , pr eventing its cyclic inter nalization.
34
mutant kap121kap123 (Gar mendia-Tor r es et al., 2007), w hile its expor t depends on the nuclear expor tin Msn5 (Gor ner et al., 2002).
The NLS PKA-consensus sequences containing the phosphor ylated Msn2 ser ine r esidue S582 is also consensus phosphor ylation by the Snf1 kinase, the or tholog of t he mammali an AMP-activated pr otein kinase in Sacchar omyces cer evisiae. In fact, Snf1 dir ectly phosphor ylates this r esidue upon pr olonged glucose depletion (De Wever et al., 2005). So, w hilst PKA contr ols Msn2 activity in glucose-gr ow ing cells, phosphor ylating at least the r esidues S582 and S620, Snf1 w ould be r equir ed to r egulate Msn2 activity in the absence of glucose by phosphor ylating the r esidue S582, allow ing posterior PKA-dependent phosphor ylation of S620 by and the consequent adaptation of the cells to glucose star vation (Mayor domo et al., 2002; De Wever et al., 2005). Snf1 is par t of a complex that is essential to many differ ent cell functions, including r egulation of the tr anscr iptional changes associated with glucose der epr ession; phosphor ylation of histone H3; dir ect r egulation of RNA polymer ase II holoenzyme; r egulation of tr anslation, glycogen biosynthesis, and lipid biosynthesis; and r egulation of gener al str ess r esponses, r esponse to salt str ess and r esponse to heat str ess (Kuchin et al., 2000; Lo et al., 2001; Alepuz et al., 1997; Ashe et al., 2000; Ber tr am et al., 2002). Ther efor e, Msn2 dow nr egulation by Snf1 phosphor ylation w ould allow yeast cells to bypass gr owth ar r est and hypothetically to coor dinate the ESR w ith the differ ent cells function in w hich Snf1 is involved.
35
Msn2 in the nucleus and cell adaptation (Dur chschlag et al., 2004; Lallet
et al., 2004; Bose et al., 2005). Ther e ar e also differ ent phosphatases that ar e able to dephosphor ylate Msn2, playing an impor tant r ole in its r egulation, antagonising PKA-dependent phosphor ylation. De Wever and collabor ator s (2005) show ed that upon a sudden glucose depletion in the medium, Msn2 activation w as dr iven by a fast but tr ansient decr ease in phosphor ylation of sever al r esidues in the NLS and this decr ease in Msn2 phosphor ylation state w as mediated by the PP1 pr otein phosphatase (encoded by GLC7), making it a potential mediator of glucose star vation signals that tar get Msn2. Besides, Mayor domo et al. (2002) show ed that in the r eg1 mutants (Reg1 is the r egulator y subunit of the Reg1/ Glc7 pr otein phosphatase complex), the r egulation of Msn2 distr ibution w as lost and the TF w as constitutively pr esent in the cytosol. Mor eover , they show ed that this behaviour w as associated to the pr esence of an abnor mal active Snf1 pr otein kinase that inhibits the nuclear localization of Msn2 upon car bon star vation.
36
I.3.2 The Heat shock r esponse: Hsf1 and Msn2/ 4
The r esponse to heat shock involves a fast induction of a conser ved gr oup of heat shock pr oteins (HSPs). In S. cer evisiae, this r esponse is mediated by the heat shock tr anscr iption factor (Hsf1) and Msn2/ 4. Hsf1 is an essential and modular pr otein highly conser ved fr om yeast to humans, consisting of a centr al cor e w ith a winged helix-tur n-helix class of DNA-binding domain, a leucine zipper domain, containing tw o
Figure 1.8: Overview of M sn2 activity via phosphorylation dependent sub-cellular localization.
37
hydr ophobic oligomer ization r epeats r equir ed for homo-tr imer ization, and a C-ter minal tr ans-activation domain (CAD) (Fig. 1.9).
HSFs bind a cis-element for med by multiple inver ted r epeats of nGAAn, designated as heat shock element (HSE), in the pr omoter s of its tar get genes, activating their tr anscr iption. Sakur ai and Takemor i (2007) identified the HSEs in 59 of 62 tar get genes of Sacchar omyces cer evisiae
Hsf1. They ver ified that the Hsf1 pr otein r ecognizes continuous and discontinuous r epeats of the nGAAn and the existence of some diver gence in the functional HSEs. Mor eover , w or k fr om Sakur ai gr oup (Hashikaw a et al., 2007) show ed that sequences of four or mor e HSEs ar e bound cooper atively by tw o HSF tr imer s. How ever , ther e ar e major differ ences in the number and r egulation of HSFs from yeasts to higher eukar yotes. Concer ning to the number of HSFs, w hile the yeasts S. cer evisiae and Schizosacchar omyces pombe have only one essential HSF, the nonver tebr ate metazoans such as the fr uit fly Dr osophila melanogast er have a single HSF that is essential for sever al impor tant cellular functions, such as oogenesis, ear ly lar val development and
38
sur vival in r esponse to sever e str ess challenges, but is dispensable for gr ow th and viability under nonstr essful conditions in adult flies (Pir kkala et al., 2001 and r efer ences ther ein). Ver tebr ate metazoan chickens and mammals, how ever , expr ess four r elated HSF genes w ith differ ent functions: Hsf1, Hsf2 and Hsf4 ar e pr esent in mammalian cells w hile Hsf3 is an avian-specific factor (Voellmy, 2004). Finally,
Ar abidopsis t haliana has tw enty one HSF genes tightly r egulated into a netw or k of inter acting pr oteins (Nover et al., 2001; Baniw al et al., 2004). It is thought that plants’ diver sity of HSF can be r elated to the constant and often sever e envir onmental aggr essions, these immotile or ganisms have to cope w ith.
Regar ding HSF r egulation fr om yeasts to high metazoan, the major differ ence in the conser ved Hsf1 r egulation is r elated to its active tr imer ization for m. In fact, w hile S. cer evisiae is able to discr iminate differ ent str ess conditions and activate the expr ession of genes r equir ed to each r esponse thr ough a single and essential Hsf1, the thr ee heat shock factor s pr esent in metazoan cells ar e activated only under specific developmental or envir onmental conditions. In the fir st case, Hsf1 for ms a tr imer complex and binds DNA constitutively in the HSEs (Pir kkala et al., 2001 and r efer ences ther ein), even in the absence of any heat shock, leading in some cases to the moder ate expr ession of heat shock genes dur ing nor mal gr ow th conditions (Liu et al., 1999). This implies that Hsf1 DNA-binding ability and tr ansactivation competence ar e r egulated independently (Voellmy, 2004) and ther e must be additional stimulus upon heat shock in or der to activate tr anscr iption. The signal tr igger ing the tr anscr iption of the tar get genes is most pr obably the str ess-induced hyper phosphorylation of Hsf1. The evidence for this assumption became clear fr om a study w ith
39
Hsf1. In this case, Hsf1 w as unable to bind a specific subset of tar get genes containing thr ee HSEs units and w as not extensively phosphor ylated in r esponse to str ess, being unable to activate genes containing this type of HSE (Hashikaw a et al., 2007). These r esults also suggest that oligomer ization is a pr er equisite for str ess-induced hyper phosphor ylation of Hsf1.
Ther e is another featur e that w as not evolutionar y conser ved and contr ibutes for the differ ential r egulation of the Hsf1 fr om yeasts to higher eukar yotes. Wher eas the metazoan HSF has only one C-ter minus tr ans-activation domain (CAD), HSFs fr om Sacchar omyces cer evisiae
and Kluver omyces lact is shar e an additional N-ter minal tr ans-activation domain (NAD) (Tr ott and Mor ano, 2003 and r efer ences ther ein), that, together w ith the CAD, ar e thought to mediate tempor al aspects of the heat shock r esponse (Chen et al., 2002). The heat shock r esponse can be divided into tr ansient and sustained r esponse, char acter ized by the incr ease in temper atur e over 35°C for less than 1h and by a pr olonged time of cell gr ow ing under higher temper atur es, r espectively. The Hsf1 NAD (fir st 65 amino acids) is thought to mediate the tr ansient r esponse, w her eas the CAD (betw een r esidues 595 and 783) is thought to be r esponsible for the sustained r esponse to stress. Mor eover , each tr ans-activation domain seems to r egulate the expr ession of specific genes. Hsf1 CAD is cr itical for the heat-induced expr ession of CUP1,
HSP82, and HSP26, w her eas its loss has no effect on the heat -induced expr ession of SSA1, SSA3, and HSP104 (Pir kkala et al., 2001 and r efer ences ther ein).
40
subsequently acquir es tr anscr iptional activity (r eview ed in Holmber g
et al., 2002). It is thought that under non-str ess conditions, Hsf1 fr om higher eukar yotic cells is maintained in its monomer ic and inactive for m thr ough the binding of Hsps (heat shock pr oteins). Upon activation, Hsf1 tr imer can be inactivated by binding of inhibitor s as the heat shock factor binding pr otein 1 (Hsbp1). Hsbp1 inter acts w ith the oligomer ization domain of an active Hsf1, ther eby negatively affecting HSF1 DNA binding activity (Satyal et al., 1998). The car boxyl-ter minal heptad r epeat (HR-C) should be involved in the suppr ession of Hsf1 tr imer ization as this domain is w ell conser ved among the ver tebr ate HSFs but poor ly conser ved in plants and S. cer evisiae HSFs, w hat could cause the constitutive tr imer ization of HSF in S. cer evisiae. Mammalian Hsf1 r egulation is fur ther r egulated at multiple levels, involving the oligomer ic status of Hsf1 and its DNA-binding ability, phosphor ylation in differ ent ser ine r esidues (r egulating Hsf1 tr anscr iptional activity both positively and negatively) and sumoylation, tr anscr iptional competence, nuclear localization or inter actions w ith r egulator y cofactor s or other tr anscr iption factor s (r eview ed in Holmber g et al., 2002; Voellmy, 2004).
Hsf1 function is not r estr icted to the heat shock r esponse. It w as alr eady show n that, among other effects, Hsf1 also mediates pr otection to the heavy metals copper and cadmium, as w ell as to oxidative str ess, thr ough the activation of the CUP1 gene (Silar et al., 1991; Sew ell et al.,
41
chr omatin in vivo. How ever , it is not yet know n if Hsf1 is a dir ect tar get of Snf1.
The HSF is also thought to be contr olled by the PKA pathw ay. Differ ent evidences place Hsf1 dow nstr eam of PKA and suggest that PKA might be involved in negative r egulation of Hsf1 activity (Yamamoto et al., 2007). How ever , it seems that only a subset of HSE-containing genes r egulated by Hsf1 is negatively r egulated by PKA, as for example the genes involved in the Hsf1-dependent r esponse to tr eatment w ith menadione, diamide, KO2, or 1-chlor o-2,4-dinitr obenzene (Fer guson et
al., 2005 and Yamamoto et al., 2007). Additionally, Hsf1 does not seem to be a dir ect tar get of PKA, as deletion of PKA causes an incr ease in Hsf1 phosphor ylation (Fer guson et al., 2005).
The second r egulator y system of the heat shock r esponse, Msn2 and Msn4, is not evolutionar y conser ved betw een yeasts and humans. These tr anscr iption factor s only exist in yeast and ar e found in the pr omoter s of most HSPs (Estr uch, 2000). Ther e is ther efor e some degr ee of cr osstalk betw een the tw o systems in or der to pr ecisely r egulate the yeast r esponse to heat and other str ess r esponse in w hich Hsf1 and Msn2/ 4 ar e involved. In this w ay, ther e ar e genes that ar e r egulated by only one of these systems, w hile other s, such as HSP26 and
42
et al. (2008) descr ibes that Hsf1 is actively involved in chr omatin r emodelling events at gene pr omoter s, w hile the r ole of the Msn2/ 4 system in these pr ocesses is poor ly under stood. Related to this the data fr om Yamamoto et al. (2008) suggest that Msn2/ 4 play a r ole befor e the exposur e to high temper atur es since, unlike Hsf1, they w er e not involved in the upr egulation of genes necessar y for the r ecover y per iod follow ing sever e heat shock and also w er e dispensable for cell gr ow th dur ing that per iod. Ther efor e Hsf1 and Msn2/ 4 w ould act differ entially befor e and after exposur e to extr eme temper at ur es to ensur e cell sur vival and gr ow th.
As Hsf1 and Msn2/ 4 ar e both r egulating the heat shock r esponse, although in differ ent aspects of yeast adaptation, w e w ould expect the existence of common r egulator s able to contr ol their activity. That is the case of the multi-functional E3 ubiquitin ligase Rsp5. In a r ecent r epor t, Haitani and Takagi (2008) show ed that Rsp5 is involved in the r epair system of str ess-induced abnor mal pr oteins and upon envir onmental str esses, pr imar ily r egulates the expr ession of Hsf1 and Msn2/ 4 at the post-tr anscriptional level, mediating their mRNA nuclear expor t.
43 I.3.3 The PKA pathway
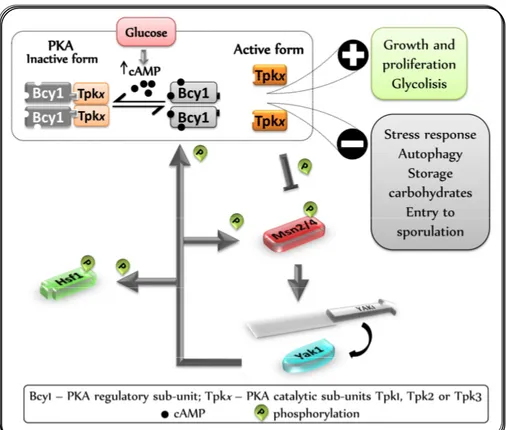
PKA (Pr otein Kinase A) pathw ay is essential for gr ow th and antagonizes induction of the gener al str ess r esponse, being also involved in many other cellular functions such as glycolysis and gluconeogenesis, aging, bud site selection, actin r epolar ization, spor ulation and pseudohyphal differ entiation (r eview ed in Santagelo, 2006). This is achieved mainly thr ough the contr ol of the phosphor ylation state of tr anscr iption activator s and r epr essor s, kinases and metabolic enzymes (Thevelein and De Winde 1999). The impor tance of PKA pathw ay for cell sur vival is r eflected in the fact that cells deficient in PKA activity stop gr ow , ar r esting in G1 and have an incr eased r esistance to heat str ess. In contr ast, cells har bour ing a constitutively activated PKA fail to ar r est in G1, ar e defective for tr ehalose and glycogen accumulation, r apidly lose viability, and r emain highly sensitive to heat str ess upon nutrient star vation (Reinder s et al., 1998). PKA, the effector kinase of this pathw ay, is thus r esponsible for coupling the nutr ients availability w ith the str ess conditions, allow ing r apid gr ow th of glucose-gr ow ing cells but simultaneously r ender ing them sensitive to envir onmental str esses to prevent cell damage (Gr iffioen and Thevelein, 2002).
PKA r egulation is quite complex, involving sever al aspects as localization of its subunits, phosphor ylation, cAMP levels, among other s, and is far beyond the scope of this r epor t. Her e I w ill give a simple over view of some PKA r egulation featur es.
44
45
to clar ify this feedback mechanism. Accor ding to these author s, Yak1 may be the effector of the PKA contr ol over the GSR, as it is able to activate both Hsf1 and Msn2 thr ough phosphorylation w hen PKA activity is low er ed by glucose depletion. An additional level of contr ol of this mechanism w as suggested by Mor iya et al. (2001) that pr ovide evidences that glucose levels may modulate the intr acellular localization of Yak1.