Cryptic diversity in
rodents from Costa
Rica.
Sara María Cáceres Valdés
Master’s in Biodiversity, Genetics and Evolution Department of Biology
2018/2019
Supervisor
Joana Maria Jorge Pereira de Castro Paupério, Postdoctoral Researcher, CIBIO-InBIO
Co-supervisor
Paulo Célio Pereira Martins Alves, Associate Professor, Faculty of Sciences, Univeristy of Porto, CIBIO-InBIO
Todas as correções determinadas pelo júri, e só essas, foram efetuadas. O Presidente do Júri,
FCUP Cryptic diversity in rodents from Costa Rica
i
Acknowledgements
I would like to thank my wonderful supervisors for making this project being possible. A huge thanks to Paulo Célio, for welcome me in Portugal with open arms from the very beginning and for giving me the chance to be part of this interesting project where he co-supervised me and gave me a lot of support throughout the process. A very special thanks goes also to my supervisor Joana Paupério, for being so patient, for giving me great guidance and helping me when I needed the most. I feel so grateful to have worked with both.
To the CTM laboratory team, Maria and Patrícia, a big thank for sharing their valued knowledge in the laboratory field, for all patience and help provided. And thanks to Sofía for not have ‘hanged’ me when an important sample flew out of the tube with alcohol because of me during my training.
To my extraordinary classmates, thank you for all the meetings, typical meals, trips on the outskirts of Porto and all the good times we shared together, that made worth it to crossing over the Atlantic Ocean to spend this last 2 years. To Alexander Lépiz, for your valuable friendship in the short time we interact and to Sara Sampaio, for your kindness, support and availability to help me.
A very emotional thanks to my beloved Panamanian friends, Darwin, Anette and Estibali, without whom this adventure would not have been so epic. Also, my dear Vítor and his wonderful family that make me feel at home and helped me feel a bit less homesick at times.
My main and deepest thank goes to my MOTHER, without her support I would never have gotten where I am. Also, to my big sister Vlasty that together with mum form an important pillar of love and strength in my life (not forgetting our Benji). I also want to thank my father, for always supporting me as much as possible. I love you all Family!
Abstract
Mesoamerica is one of the most important biodiversity hotspots, sheltering high levels of endemism and species diversity. This region holds about 6% of the World’s mammal diversity, of which, approximately 180 species are rodents.
Within Mesoamerica, Costa Rica, is considered one of the 25 most biodiverse countries on the planet with the highest number of species per unit area. This country serves as corridor for diverse groups of mammals from the north to the south of the American continent. Therefore, the exchange of species between both regions, in addition to its topogeographical features, may have favoured crypic genetic diversity in some rodent groups as recent molecular studies and records of new species suggest.
Considering the limited knownledge on the diversity of small mammals in Costa Rica and the importance of cryptic species for biodiversity conservation, this study aimed to analyse the spatial patterns of genetic variability in some rodent species, where high diversity was previously detected.
Hence, 221 tissue samples from four genera of Cricetidae and one genus of Echimyidae were analysed. Of these, 15 were museum samples from the National Museum of Costa Rica and the Zoology Museum of the National University of Costa Rica. The genetic diversity was determined by amplifying two mitochondrial (cytb and COI) and two nuclear (IRBP and RAG1) genes.
Our results show high genetic diversity in all the genera analysed, congruent with the geographic distribution of the species in Costa Rica. Some of the recognised lineages show deep divergence, and may even correspond to new cryptic species.
This study describes cryptic diversity in small mammals from Costa Rica, highlighting the importance of this country for the conservation of biodiversity in Mesoamerica.
FCUP Cryptic diversity in rodents from Costa Rica
iii
Keywords
Small mammals, cryptic species, mitochondrial DNA, nuclear DNA, genetic diversity, Mesoamerica.
Resumo
A Mesoamérica é uma das regiões do mundo com maior biodiversidade, albergando elevados níveis de endemismo e diversidade de espécies. Esta região detém cerca de 6% da diversidade de mamíferos ao nível mundial, incluindo aproximadamente 180 espécies de roedores.
Dentro da Mesoamérica, a Costa Rica é considerada um dos 25 países com maior biodiversidade do planeta, com o maior número de espécies por unidade de área. Este país constitui um corredor para diversos grupos de mamíferos do norte ao sul do continente americano. Deste modo, o intercambio de espécies entre as duas regiões, assim como as suas características topogeográficas, pode ter favorecido a diversidade genética críptica em alguns grupos de roedores, como sugerem alguns estudos moleculares e descrições recentes de novas espécies.
Considerando a escassez de conhecimento relativo à diversidade de pequenos mamíferos na Costa Rica e a importância da detecção de espécies crípticas para a conservação da biodiversidade, este estudo teve como objetivo principal o estudo dos padrões espaciais da variabilidade genética de alguns géneros das famílias Cricetidae e Echimyidae, nos quais tinha sido detectada recentemente elevada diversidade.
Assim, neste estudo foram analisadas 221 amostras de tecido de quatro géneros de Cricetidae e de um género de Echimyidae. Destas, 15 eram do Museu Nacional da Costa Rica e do Museu de Zoologia da Universidade Nacional da Costa Rica. Foram amplificados e sequenciados dois genes mitocondriais (cytb e COI) e dois genes nucleares (IRBP e RAG1), com o objetivo de analisar a diversidade genética desses géneros na região.
Os resultados obtidos permitiram detectar uma elevada diversidade genética em todos os géneros analisados, concordantes com a distribuição geográfica das espécies na Costa Rica. Algumas das linhagens identificadas apresentam uma elevada divergência podendo eventualmente corresponder a espécies crípticas.
Este estudo descreve a ocorrência de diversidade críptica em pequenos mamíferos na Costa Rica, salientado a importância deste país para a preservação da biodiversidade na Mesoamérica.
FCUP Cryptic diversity in rodents from Costa Rica
v
Palavras chave
Pequenos mamíferos, espécies crípticas, ADN mitocondrial, ADN nuclear, diversidade genética, Mesoamérica.
Table of Contents
Acknowledgements ... i Abstract ... ii Keywords ... iii Resumo... iv Palavras chave ... v Table of Contents ... viList of Tables ... vii
List of Figures ... x
1. Introduction ... 1
1.1. Costa Rica, a biodiversity hotspot ... 1
1.2. New World rodent cryptic speciation ... 3
1.2.1. Genus Oligoryzomys ... 5 1.2.2. Genus Nyctomys... 6 1.2.3. Genus Scotinomys ... 7 1.2.4. Genus Reithrodontomys ... 8 1.2.5. Genus Proechimys ... 10 1.3. Molecular markers ... 11 1.4. Objectives... 13
2. Material and Methods ... 14
2.1. Study area and sampling ... 14
2.2. DNA extraction, amplification and sequencing ... 15
2.2.1. Cytochrome b (cytb) ... 15
2.2.2. Cytochrome c oxidase subunit I (COI) ... 16
2.2.3. IRBP and RAG1 nuclear genes ... 18
2.3. Phylogenetic analyses... 19 3. Results ... 21 3.1. Phylogenetic analyses... 21 3.1.1. Genus Oligoryzomys ... 21 3.1.2. Genus Nyctomys... 26 3.1.3. Genus Scotinomys ... 30 3.1.4. Genus Reithrodontomys ... 34 3.1.5. Genus Proechimys ... 39 5. References ... 46 6. Supplementary material ... 60
FCUP Cryptic diversity in rodents from Costa Rica
vii
List of Tables
Table 1. List of rodents of the families Cricetidae and Echimyidae that currently occur
in Costa Rica and surrounding area, according to Rodríguez-Hernández et al. (2014).
Pg.5 Table 2. Genera, number of species and analysed samples using mitochondrial (Cytb
and COI) and nuclear (RAG1 and IRBP) genes, with reference to the sampling sites
Pg.14 Table 3. Primers used for amplification and sequencing of Cytb gene. Pg.16 Table 4. Primers pair and annealing temperature optimized in each genus for Cytb
gene amplification.
Pg.16 Table 5. Primers used for amplification and sequencing COI fragments B2 and LCn. Pg.17 Table 6. Primers used for amplification and sequencing COI fragment SFF. Pg.17 Table 7. Primers used for amplification and sequencing of IRBP and RAG1 genes
respectively.
Pg.18 Table 8. Primers pair and annealing temperature optimized in each genus for IRBP and
RAG1 genes amplification.
Pg.19 Table 9. Genetic indices for intraspecific diversity of the genus Oligoryzomys with COI
mtDNA sequences (657 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: number of haplotypes; S: polimorphic sites; Hd: Haplotype diversity; π: nucleotide diversity.
Pg.22
Table 10. Estimates of interspecific diversity between clades of the genus Oligoryzomys
in the lower triangle for COI (657 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.22 Table 11. Genetic indices for intraspecific diversity of the genus Oligoryzomys with IRBP
nDNA sequences (541 bp) for each clade identified according to the Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.22
Table 12. Estimates of interspecific diversity between clades of the genus Oligoryzomys
in the lower triangle for IRBP sequences (541bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.23 Table 13. Genetic indices for intraspecific diversity of the genus Oligoryzomys with
RAG1 nDNA sequences (952 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: haplotype diversity; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.23
Table 14. Estimates of interspecific diversity between clades of the genus Oligoryzomys
in the lower triangle for RAG1 sequences (952 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.23 Table 15. Genetic indices for intraspecific diversity of the genus Nyctomys with cytb
mtDNA sequences (1140 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: haplotype diversity; S: polimorphic sites; Hd: Haplotype diversity; π: nucleotide diversity.
Table 16. Estimates of interspecific diversity between clades of the genus Nyctomys and
outgroups in the lower triangle for cytb sequences (1040 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.27 Table 17. Genetic indices for intraspecific diversity of the genus Nyctomys with COI
mtDNA sequences (657 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: haplotype diversity; S: polimorphic sites; Hd: Haplotype diversity; π: nucleotide diversity.
Pg.27
Table 18. Estimates of interspecific diversity between clades of the genus Nyctomys and
outgroups in the lower triangle for COI sequences (657 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.27 Table 19. Genetic indices for intraspecific diversity of the genus Nyctomys with IRBP
nDNA sequences (912 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.28
Table 20. Estimates of interspecific diversity between clades of the genus Nyctomys in
the lower triangle for IRBP sequences (921 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.28 Table 21. Genetic indices for intraspecific diversity of the genus Scotinomys with cytb
mtDNA sequences (1140 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: number of haplotypes; S: polimorphic sites; Hd: Haplotype diversity; π: nucleotide diversity.
Pg.31
Table 22. Estimates of interspecific diversity between clades of the genus Scotinomys in
the lower triangle for cytb sequences (1140 bp). Upper triangle shows the standard error from 500 bootstrap.
Pg.31 Table 23. Genetic indices for intraspecific diversity of the genus Scotinomys with COI
mtDNA sequences (657 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: number of haplotypes; S: polimorphic sites; Hd: Haplotype diversity; π: nucleotide diversity.
Pg. 31
Table 24. Estimates of interspecific diversity between clades of the genus Scotinomys in
the lower triangle for COI sequences (657 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg. 31 Table 25. Genetic indices for intraspecific diversity of the genus Scotinomys with RAG1
nDNA sequences (779 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg. 31
Table 26. Estimates of interspecific diversity between clades of the genus Scotinomys in
the lower triangle for RAG1 (779 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg. 32 Table 27. Genetic indices for intraspecific diversity of the genus Reithrodontomys with
cytb mtDNA sequences (1140 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
FCUP Cryptic diversity in rodents from Costa Rica
ix
Table 28. Estimates of interspecific diversity between clades of the genus
Reithrodontomys in the lower triangle for cytb sequences (1140 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg. 35 Table 29. Genetic indices for intraspecific diversity of the genus Reithrodontomys with
COI mtDNA sequences (657 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.35
Table 30. Estimates of interspecific diversity between clades of the genus
Reithrodontomys in the lower triangle for COI sequences (657 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.35 Table 31. Genetic indices for intraspecific diversity of the genus Reithrodontomys with
RAG1 mtDNA sequences (1253 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.35
Table 32. Estimates of interspecific diversity between clades of the genus
Reithrodontomys in the lower triangle for RAG1 sequences (1253 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.36 Table 33. Genetic indices for intraspecific diversity of the genus Proechimys with cytb
mtDNA sequences (1127 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.39
Table 34. Estimates of interspecific diversity between clades of the genus Proechimys
in the lower triangle for cytb sequences (1127 bp). Upper triangle shows the standard error from 500 bootstrap replicates.
Pg.40 Table 35. Genetic indices for intraspecific diversity of the genus Proechimys with COI
mtDNA sequences (657 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.40
Table 36. Estimates of interspecific diversity between clades of the genus Proechimys
in the lower triangle for COI sequences (657 bp). Upper triangle shows the standard error from 500 bootstrap.
Pg.40 Table 37. Genetic indices for intraspecific diversity of the genus Proechimys with RAG1
mtDNA sequences (918 bp) for each clade identified according to Bayesian inference. N: Number of sequences obtained in this study (retrieved sequences from GenBank in parentheses); H: Number of haplotypes; S: Polymorphic sites; Hd: Haplotype diversity; π: Nucleotide diversity.
Pg.40
Table 38. Estimates of interspecific diversity between clades of the genus Proechimys
in the lower triangle for RAG1 sequences (918 bp). Upper triangle shows the standard error from 500 bootstrap.
Pg.40 Table S1. Information of both mitochondrial and nuclear DNA sequences retrieved from
GenBank and BOLD Systems for this research.
Pg.60 Table S2. Information of small mammals sampled in Costa Rica. Pg.71
List of Figures
Figure 1. Mesoamerican region according to Olguín-Monroy et al. (2013) pg.1 Figure 2. Distribution of the genus Oligoryzomys (obtained from https://www.gbif.org/) pg.6 Figure 3. Distribution of the genus Nyctomys, with a detailed view in Mesoamerica
(obtained from https://www.gbif.org/)
pg.7 Figure 4. Distribution of the genus Scotinomys, with a detailed view of Mesoamerica
(obtained from https://www.gbif.org/)
pg.8 Figure 5. Grouping of recognized species of the genus Reithrodontomys per subgenus; * =
Species found in Costa Rica
pg.9 Figure 6. Distribution of the genus Reithrodontomys (obtained from https://www.gbif.org/) pg.9 Figure 7. Distribution of the genus Proechimys (obtained from https://www.gbif.org/) pg.10 Figure 8. Map of Costa Rica depicting the collecting localities of different samples of
Cricetidae and Echimyidae available for analyses.
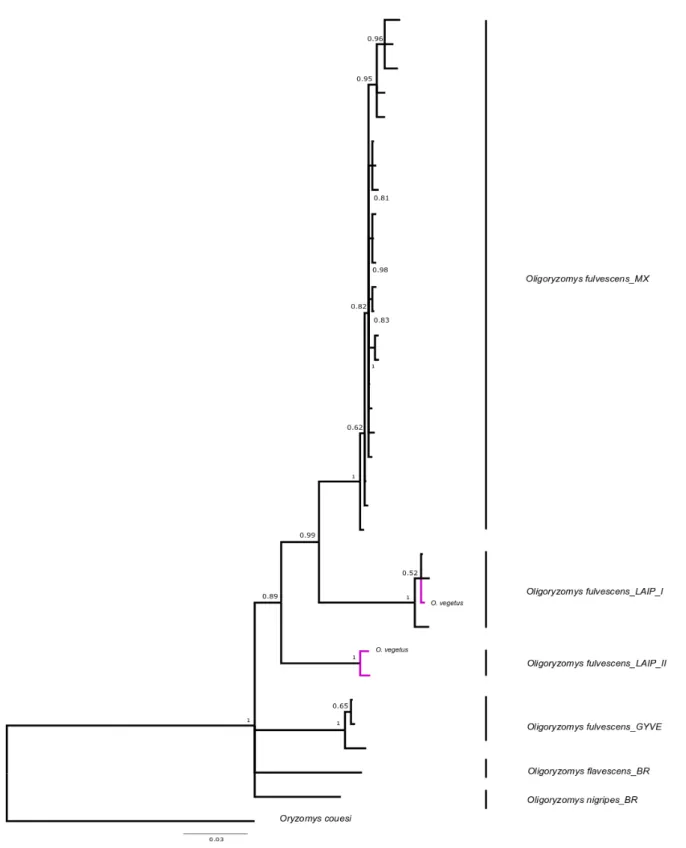
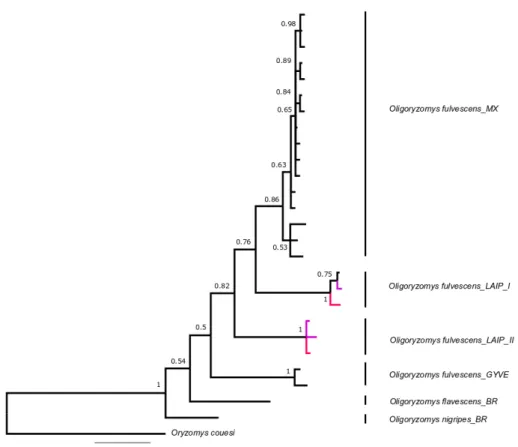
pg.14 Figure 9. COI Primer sets for DNA barcoding of small mammals from Costa Rica. pg.17 Figure 10. Bayesian inference tree for genus Oligoryzomys for COI gene (657 bp). Purple
branches represent our samples. Probabilities of major nodes are indicated. MX= Mexico; LAIP= La Amistad International Park, Costa Rica; GYVE= Guiana-Venezuela; BR= Brazil. Roman numbers represent the number of the clade. Outgroup: Oryzomys couesi.
pg.24
Figure 11. Bayesian inference tree for genus Oligoryzomys for COI gene (420 bp). Purple branches represent our samples and museum samples are shown in red. Probabilities of major nodes are indicated. MX= Mexico; LAIP= La Amistad International Park, Costa Rica; GYVE= Guiana-Venezuela; BR= Brazil. Roman numbers represent the number of the clade. Outgroup: Oryzomys couesi.
pg.25
Figure 12. Gene haplotypes network from the genus Oligoryzomys. A) IRBP (541 bp), B) IRBP (952 bp) and C) RAG1 (952bp). Squares represent different geographical regions.
pg.25 Figure 13. Bayesian inference tree for genus Nyctomys for cytb gene (1143 bp). Purple
branches represent our samples. Probabilities of major nodes are indicated. ESHN= El Salvador-Honduras; LAIP= La Amistad International Park, Costa Rica; SV= Selva Verde, Costa Rica; GU= Guatemala; MX= Mexico. Roman numbers represent the number of the clade. Outgroup: Tylomys nudicaudus
pg.28
Figure 14. Bayesian inference tree for genus Nyctomys for COI gene (657 bp). Purple branches represent our samples and museum samples in red. Probabilities of major nodes are indicated. Were: ESGU= El Salvador-Guatemala; MVCR= Monteverde, Costa Rica; SV= Selva Verde, Costa Rica; GU= Guatemala. Roman numbers represent the number of the clade. Outgroup: Tylomys nudicaudus .
pg.29
Figure 15. IRBP (912 bp) haplotype network from the genus Nyctomys. pg.29 Figure 16. Bayesian inference tree for genus Scotinomys for cytb gene (1140 bp). Purple
branches represent our samples and museum samples are in red. Probabilities of major nodes are indicated. HN= Honduras; BCNP= Braulio Carrillo National Park, Costa Rica; LAIP= La Amistad International Park; CVI= Cartago Volcán Irazú, Costa Rica. Roman numbers represent the number of the clade. Outgroup: Reithrodontomys creper.
FCUP Cryptic diversity in rodents from Costa Rica
xi
Figure 17. Bayesian inference tree for genus Scotinomys for COI gene (657 bp). Purple branches represent our samples. Probabilities of major nodes are indicated. LAIP=La Amistad International Park, Costa Rica; CVI= Cartago, Volcán Irazú, Costa Rica; BCNP= Braulio Carrilo National Park; HN= Honduras; CR= Costa Rica; ESGUNIHN= El Salvador-Guatemala-Nicaragua-Honduras; SAES= Santa Ana, El Salvador; MX= Mexico. Roman numbers represent the number of clade. Outgroup: Reithrodontomys creper.
pg.33
Figure 18. RAG1 (779bp) haplotype network from the genus Scotinomys. pg.33 Figure 19. Bayesian inference tree for genus Reithrodontomys for cytb gene (1140 bp).
Purple branches represent our samples and museum samples are in red. LAIP= La Amistad International Park, Costa Rica; CVI= Cartago, Volcán Irazú, Costa Rica; BCNP= Braulio Carrilo National Park Probabilities of major nodes are indicated. Outgroup: Peromyscus mexicanus.
pg.36
Figure 20. Bayesian inference tree for genus Reithrodontomys for COI gene (658 bp). Purple branches represent our samples and museum samples are in red. LAIP= La Amistad International Park, Costa Rica; CRNIES= Costa Rica-Nicaragua-El Salvador; CRMXGU= Costa Rica-Mexico-Guatemala; BCNP= Braulio Carrilo National Park; MXUSA= Mexico-United States; MX=Mexico; GU= Guatemala; USA= United States; NA= Not available. Probabilities of major nodes are indicated. Outgroup: Scotinomys teguina.
pg.37
Figure 21. A) RAG1 (1253 bp) and B) IRBP (894bp) haplotype networks from the genus Reithrodontomys.
pg.38 Figure 22. Bayesian inference tree for genus Proechimys for COI gene (658 bp). Purple
branches represent our samples and museum samples are in red. Probabilities of major nodes are indicated. Outgroup: Hoplomys gymnurus.
pg.41 Figure 23. Bayesian inference tree for genus Proechimys for cytb gene (1127 bp). Purple
branches represent our samples and museum samples are in red. Probabilities of major nodes are indicated. Outgroup: Hoplomys gymnurus.
pg.42 Figure 24. RAG1 (918 bp) haplotype network from the genus Proechimys. pg.42
List of Abbreviations
ALTMB Altamira, Amazonas, Brazil
AMV Amazonas, Venezuela
AR Argentina
ATH Atlántica, Lancetilla Botanical Garden, Honduras
BBWG Baramita, Barima-Waini, Guiana
BDC Base do Carmo, Sao Paulo, Brazil
BLP Boweron, La Lomita, Paraguay
BOLD Barcode of Life Data System
BPBC San Jose, Escazu, Base Of Pico Blanco, Costa Rica
BR Brazil
BVGU Baja Verapaz, Guatemala
CB Cerrado, Brazil
CCC Ciuddad Colón, Costa Rica
CCHR Cerro Chirripo, Costa Rica
CHM Chiapas, México
CJAB Cadeias do Jamari, Amazon, Brazil
CLCR Cartago, La Carpentera, Costa Rica
CMB Cochambamba, Bolivia
COI Cytochrome c Oxidase I
CPCM Campeche, Mexico
CPCR Estación Biológica Caño Palma, Tortuguero, Costa Rica
CR Costa Rica
CRT Cartago Proince, Costa Rica
CSAG Chimaltenango, Santa Apolonia, Guatemala
CSTC Cartago, Santa Cruz, Costa Rica
CVI Cartago, Volcán Irazú, Costa Rica
Cytb Cytochrome b
DNA Deoxyribonucleic Acid
EC Ecuador
ES El Salvador
FCUP Cryptic diversity in rodents from Costa Rica
xiii
FGY French Guiana
FVL Finca Vuelta Larga, Sucre, Venezuela
GBIF Global Biodiversity Information Facility
GDH Gracias a Dios, Honduras
GU Guatemala
GY Guiana
HCR 2 km N, 0.5 km E de Sacramento, Heredia, Costa Rica
HN Honduras
IRBP Interphotoreceptor Retinoid Binding Protein
JAB Jainu, Amazonas, Brazil
JCBNP Alajuela; Parque Nacionale Juan Castro Blanco, Costa Rica
LLCR Iintesección con La Lucha, Costa Rica
LNG Les Nouragues, French Guiana
MGB Minas Gerais, Brazil
MVCR Monteverde, Costa Rica
MX Mexico
NA Not available
Nam North America
NCBI National Center of Biotechnology Information
NI Nicaragua
OCH Olancho, Catacamas, Honduras
OICM Ometepe Island, Cerro Madera, NICARAGUA
OUS Oklahoma, USA
PACR Puntarenas, Costa Rica
PARF Pampa-Atlantic Rain Forest
PCD Portuguesa, Cano Delgadito, Venezuela
PCOA Chiriqui, Ojo De Agua, Panama
PCR Polymerase Chain Reaction
PGU Petén, Guatemala
PM Distrito Federal, Parres, Mexico
PMG Pic Matecho, French Guiana
PNAMH Comayagua, Parque Nacional Azul Meámbar, Honduras PNMES National Park Montecristo, El Salvador
PTVE Pichincha, Tandayapa Valley, Ecuador
QRSM Quintana Roo, San Miguel, MEXICO
RAG1 Recombination Activating Gene 1
RN Río Negro, Argentina
RNI Rivas, Nicaragua
ROVC Reserva Oro Verde, San Josecito, Costa Rica
RVNP Quebrada Provición, Parque Nacional Rincón de la Vieja, Costa Rica
SAES Santa Ana, El Salvador
SCGU Sacatepequez, Guatemala
SEPA Santiago del Estero, Pellegrini, Argentina
SG Saül, French Guiana
SGCR San Gerardo, Costa Rica
SIGC San Isidro del General, Costa Rica
SJ San José, Costa Rica
SJMG Saint Jean du Maroni, French Guiana
SLMMG St-Laurent-du-Maroni, Maripasoula, French Guiana SPCB Capao Bonito, Sao Paulo, Brazil
SPPB Pedreira, Sao Paulo, Brazil
SSES San Salvador, El Salvador
TBM Tabasco, Mexico
TDGB Terezina de Goias, Goias, Brazil
TLM Tamaulipas, Mexico
TMG Trinite Mountains, French Guiana
TTPB Tocantins, Tocantins,Parana, Brazil
USA United States of America
VBCR Varablanca, Costa Rica
VCM Veracruz, Mexico
VE Venezuela
VMC Villa Mills, Costa Rica
YLBM Yucatan, Laguna Becanchen, Mexico
YM Yucatan, Mexico
ZCGU Zacapa, Guatemala
FCUP Cryptic diversity in rodents from Costa Rica
1
1. Introduction
1.1. Costa Rica, a biodiversity hotspot
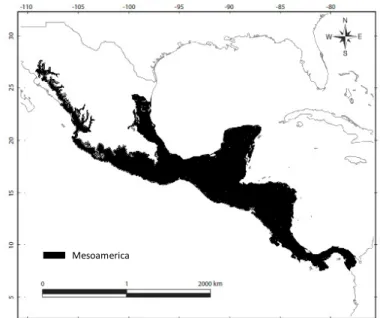
Mesoamerica, extending from south-eastern Mexico to the countries of Central America (Figure 1), is a biodiversity hotspot (Myers et al. 2000) in which the levels of endemism and species diversity are most likely underestimated. Biologically, it is one of the most complex regions of the world (Almendra & Rogers 2012) occupying the second most important hotspot after the Tropical Andes (Myers et al. 2000). Its topography and biogeographic history involve relatively recent events of extensive diversification in situ due to climatic changes and geologic processes during the Great American Biotic Interchange (Stehli & Webb 1985; Cody et al. 2010) where Central America served as a species bridge between Nearctic and Neotropical biogeographic realms (Halffter 1987; Marshall & Liebherr 2000; Morrone 2010; Webb 2006).
Figure 1. Mesoamerican region according to Olguín-Monroy et al. (2013).
The Mesoamerican region has been considered to support more than 6% of the world’s mammalian diversity (Ceballos & Ehrlich 2006; Ceballos et al. 2002; Reid 2009). The environments ranging from low elevation savanna, semiarid scrub and humid tropical forests to montane systems that exceed 4000 m in elevation (Savage 1982) promote an area with high levels of diversification sheltering nearby 440 mammal species (León-Paniagua et al. 2007). The endemism of mammals in this region is mostly owing to the vast diversity of small mammals, encompassing 20 species of shrews and more than 180 species of rodents (León-Paniagua et al. 2007).
Furthermore, roughly 100 mammal species, approximately 30% of the total, are endemic to Central America (Jenkins & Giri 2008; Reid 2009), one of the youngest subregions
of the Neotropical realm founded by the formation of the Panamanian land bridge. At the specific level, Almendra and Rogers (2012) adduce Central America supports 69 endemic rodents, in 27 genera, including Isthmomys, Nyctomys, Otonyctomys, Ototylomys, Rheomys,
Scotinomys, and Syntheosciurus. Besides, 14 bats (11 genera), 11 shrews (including 10 Cryptotis and one Sorex), four primates (two Alouatta, Ateles geoffroyi, and Saimiri oerstedii),
four carnivores (two Bassaricyon, Bassariscus sumichrasti, and Procyon pygmaeus), three lagomorphs (two Sylvilagus and Lepus flavigularis), two marsupials (Marmosa mexicana and
Marmosops invictus), and one sloth (Bradypus pygmaeus) are also endemic.
Relative to its land area, Costa Rica is internationally recognised for its abundant biodiversity (Gardner & Carleton 2009) supporting a high diversity of mammals compared to larger countries such as Mexico. This high richness is mostly attributable to its topogeographical features, the wide variety of habitats and ecosystems as well as the neotropical climate varying according to the altitudinal gradient (McCain 2004; Kluge et al. 2006), being considered the country with the highest density of biodiversity worldwide (Obando 2002). From the ecosystem point of view, cloud forests are the regions with the highest rate of endemism in Costa Rica (Obando 2002; Kohlmann et al. 2007), in this regard, endemic mammals are mostly found in this environments, mainly in the highlands. The number of rodent species herein are exceeded only by the order Chiroptera, being that, of the mammals in Costa Rica, 114 species are Chiroptera and 47 Rodentia (Rodríguez-Herrera et al. 2014).
Rodríguez-Herrera et al. (2014) recorded 11 new species within the political limits of Costa Rica, totalling 249 species in the national list of mammals, of which six are endemic, all rodents: Orthogeomys heterodus, O. cherriei, Heteromys oresterus, H. nubicolens,
Reithrodontomys rodriguezi and R. musseri. Alongside with Panama, Costa Rica has served
as a corridor and barrier for diverse groups of mammals from the north and south of the continent, giving rise to an assemblage with a mixture of species originating in both hemispheres (Cody et al. 2010; Wilson et al. 2014). Bearing in mind that fact and taking in consideration the geographical region composed by the highlands of the Talamanca mountain range in Costa Rica and the Chiriquí area of western Panama, the number of endemic mammals rises to 21 species. This number can further be increased to 23 regional endemic species if two shared species, Reithrodontomys brevirostris and R. paradoxus, in the far north of the country with Nicaragua, are considered.
FCUP Cryptic diversity in rodents from Costa Rica
3
1.2. New World rodent cryptic speciation
The members of the order Rodentia constitute the most diverse group of mammals with an approximate number of 2277 species (Musser & Carleton 2005) and corresponding to nearly 40% of the known living mammal diversity. Rodents are worldwide distributed with the exception of Antarctica, inhabiting diverse ecosystems from forests, savannahs, grasslands, mountains, deserts, rivers to swamps, displaying terrestrial, arboreal, fossorial or semi-aquatic habits. Therefore, through presenting different ecological niches, they play key roles as seed and spore dispersers, pollinators, seed predators, contributors of energy and nutrient cycling, plant succession and species composition modifiers, and as a food source for many predators (Villa-Cornejo et al. 1998; Aplin & Singleton 2003; Brakes & Smith 2005).
Taking into account this group of animals are able to adapt and they have particular characteistics such as a high reprodutive potential, it is important to considere variability between species. All these facts may have favoured the appearance of cryptic species, as is the case of some members belonging to the Cricetidae family, a highly successful group within the large and complex rodents’ superfamily Muroidea (Musser & Carleton 2005), composed by five subfamilies: Sigmodontinae, Cricetinae, Arvicolinae, Tylomyinae and Neotominae. A debate regarding systematics in this group stil remains (Miller & Engstrom 2008). Moreover, Echimyidae family is an ancient family originated in South America whose fossils date from the mid-Eocene (Antoine et al. 2012). It is one of the most diverse southamerican rodent families and, like the Cricetidae family, it presents an unresolved taxonomy due to the great morphological similarity among its members (Antoine et al. 2012).
Cryptic species is a designation of those species that are, or have been, erroneously classified as a single nominal taxon due to their morphological similarity (Bickford et al. 2007; Beheregaray & Caccone 2007). Speciation is not always matched by morphological change, consequently, the accurate number of biological species is likely to be greater than the current estimate of nominal species, most of which were defined on purely morphological information. Cryptic lineage recognition can be severely impacted by morphological stasis (Pfenninger & Schwenk 2007). Therefore, is not surprising that most recent cryptic species studies rely largely on genetic data (Beheregaray & Caccone 2007; Bickford et al. 2007).
There are several hypotheses regarding to the origin of the family Cricetidae in Mesoamerica. On one hand, based on fossil evidence, some researchers hypothesize that the members of this rodent family entered South America preceding the formation of the Panamanian bridge (Marshall 1979; Woodburne & Swisher 1995). On the other hand, others suggest that cricetids diversified in southern Central America and once the Panamanian land bridge was completed in the Pliocene (3–2.7 Ma), one or several lineages of cricetids were
among the first mammalian groups to enter South America (Pardiñas et al. 2002; Steppan et al. 2004). Recently, Verzi and Montalvo (2008) described late Miocene fossils belonging to the rodent subfamily Sigmodontinae and the carnivore family Mustelidae from the Cerro Azul Formation in Caleufú, Argentina, from an earlier time (5.8–5.7 Ma). However, Prevosti and Pardiñas (2009) argued that the age of this site is not well-established and proved that the alleged carnivore is actually a didelphimorph marsupial.
During the Pleistocene, when climatic fluctuations caused continual modifications in forest ranges and recurrent cycles of expansion and contraction of populations in a complex system of refugia, habitat-restriction among Neotropical species may have led to genetic structuring and accelerated speciation (Clare 2011). In fact, undersampled regions such as neotropical rainforests may contain a large amount of species thus far waiting to be discovered (Costello et al. 2013; Mora et al. 2011; Wheeler et al. 2012).
Nonetheless, it seems that vicariant events driven by climatic oscillations during the Pleistocene (or earlier) were enough to promote New World rodent speciation. Panama’s Darién region was likely isolated from South America until ∼13 to 7 Ma (Coates et al. 2004) and from Central America until the formation of the Panamanian land bridge. Thereby, Marshall (2007) regards the Darién as a separate physiographic province. A series of species-level splits have been identified within several rodent taxa distributed in eastern Panama, compared with populations in western Panama and Costa Rica. Although narrow in geographic sampling, molecular phylogenetic studies have uncovered a series of species-level rodent taxa in lower Central America. These include Melanomys caliginosus (Hanson & Bradley 2008), Heteromys desmarestianus, and H. australis (Rogers & González 2010). A similar pattern was also recovered for other groups of mammals, like Alouatta pigra and A.
palliate, species whose distributions are not restricted to Central America.
Among other examples, Bradley et al. (2008) determined that Sigmodon toltecus (generally distributed north of the Central American Highland Massif) and S. hirsutus (southern Central America and northern South America) were sister taxa. Hanson et al. (2010) evaluated genealogical relationships among samples of Oryzomys couesi from Mexico and Central America. Four species-level clades were identified, two from the Atlantic and Pacific versants in northern Central America respectivelly, a third from the Atlantic coast of Costa Rica, and a fourth from the Pacific coast of central Panama. Additionally, Rogers and González (2010) confirmed the species status of Heteromys nubicolens, a species known only from the Cordillera de Tilarán and Cordillera de Guanacaste, Costa Rica, and whose sister taxon, H.
oresterus, occurs to the south in the Cordillera de Talamanca, Costa Rica (Anderson & Timm
FCUP Cryptic diversity in rodents from Costa Rica
5
Currentlly, 18 Cricetidae and 2 Echimyidae small mammal genera are documented from Costa Rica (Table 1, Rodríguez-Herrera et al. 2014). Several of these are arboreal, such as Tylomys, Otolomys and Nyctomys. Aquatic mice are represented by two species of
Rheomys. Costa Rican Reithrodontomys (8 species) are mostly restricted to highland forms,
as well as Peromyscus. Scotinomys are highland mice that are active during the day and make audible vocalizations. Moreover, Proechimys is one of the most specious genus of the rodent family Echimyidae.
Table 1. List of rodents of the families Cricetidae and Echimyidae that currently occur in Costa Rica and
surrounding area, according to Rodríguez-Herrera et al. (2014).
Family Subfamily Genus # of
species
Restricted distribution
North Costa Rica
and Nicaragua Costa Rica
Costa Rica and Panamá highlands
Cricetidae
Sigmodontinae Rheomys 2 R. underwoodi
Sigmodon 1 Handleyomys 1 Melanomys 1 Nephelomys 1 N. devius Oecomys 1 Oligoryzomys 2 O. vegetus Oryzomys 1 Sigmodontomys 1 Tanyuromys 1 Transandinomys 2 Zygodontomys 1 Tylominae Nyctomys 1 Ototylomys 1 Tylomys 1
Neotominae Scotinomys 2 S. xerampelinus
Peromyscus 1 Reithrodontomys 8 R. brevirostris, R. paradoxus R. musseri, R. rodriguezi R. creper Echimyidae Proechimys 1 Hoplomys 1
1.2.1. Genus Oligoryzomys
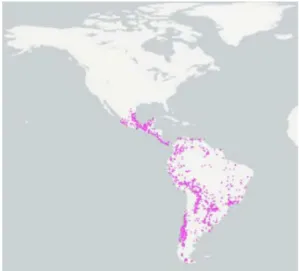
Species of the cricetid genus Oligoryzomys, commonly known as pygmy rice rats or colilargos, are found across most Neotropical biomes, and represent a relatively diverse radiation of small Neotropical sigmodontine mice, occurring from Mexico throughout Central America southward into Tierra del Fuego (Figure 2; Hall 1981; Emmons 1997). It is one of the most complex and diverse genus of the subfamily Sigmodontinae and the tribe Oryzomyini. Among mammals, it is distinguished for presenting remarkable taxonomic inconsistences, since the number of species is not a consensus among the authors. Musser and Carleton (2005) recognised 18 Oligoryzomys species, initially divided amongst five species groups (Carleton & Musser 1989). Thus, it is likely that the number of described species underestimates the actual biodiversity in this group (Musser & Carleton 2005), especially given the amount of inter- and intraspecific diversity documented in the genus by Weksler and
Bonvicino (2005). Of the 18 Oligoryzomys species currently identified, most are found exclusively in South America (Musser & Carleton 2005); only O. fulvescens and O. vegetus spread in Central and North America (Hall 1981). Carleton and Musser (1995) documented several places of sympatry between the upland Oligoryzomys vegetus and lower-elevation O.
fulvescens. Oligoryzomys vegetus is restricted to eastern Panama and Costa Rica highlands
(Carleton & Musser 1995), a region which has formed a modest center for mammalian endemism in southern Central America, generally above 1000 m elevation and within lower montane and montane biotic zones. Whereas O. fulvescens occurs in the Sierra Madre Oriental and Occidental of Mexico south and east into Central America and northern South America (Carleton & Musser 1989; Musser & Carleton 2005) from sea level to 1000 meters in wet tropical and subtropical associations. Mexican and Mesoamerican populations of O.
fulvescens are divided into 8 subspecies (Hall 1981; Carleton & Musser 1995). Fairly, not
much is known regarding genetic variation within or among northern taxa relative to their South American correlatives.
Figure 2. Distribution of the genus Oligoryzomys (obtained from https://www.gbif.org/).
1.2.2. Genus Nyctomys
Sumichrast’s vesper mice of the genus Nyctomys, from the Greek nyx = night and mys = mouse, are unusual as they are arboreal rodents, endemic to Mesoamerica. Nyctomys
sumichrasti (Cricetidae, Tylomyinae; Musser & Carleton 2005) is found from southern Jalisco
and Veracruz in Mexico to eastern Panama excluding the Yucatán Peninsula (Figure 3; Hunt et al. 2004; Pérez-Lustre & Santos-Moreno 2010; Rodríguez-Herrera et al. 2014). It inhabits evergreen lowlands and lower montane regions including cloud, secondary, riparian, and semi-deciduous forests (Hall 1981; Sánchez-Hernández et al. 1999; Cervantes et al. 2004; Hunt et al. 2004) preferring middle and upper level forest strata, ranging elevations from sea level to 1800 m (Emmons 1997; Timm & LaVal 2000; Hunt et al. 2004; Reid 2009), rarely
FCUP Cryptic diversity in rodents from Costa Rica
7
descending to the ground. It is considered as sister taxa of two other mesoamerican endemics
Tylomys and Otolomys but supposed to be closely related to Otonyctomys (Timm & LaVal
2000). These tylomyine genera are considered remnants of a clade that diverged relatively early in evolutionary history of sigmodontine rodents (Carleton 1980; Steppan 1995). Considering intraspecific variation, this diverse taxon encompasses approximately nine subspecies acknowledged by Hall (1981): colimensis, costaricensis, decolorus, florencei,
nitellinus, pallidulus, salvini, sumichrasti, and venustulus. However, little has been reported
about the taxonomy, geographic and genetic variation, or relationships between them. Musser and Carleton (2005) argue the existence of two groups corresponding to Nyctomys populations occurring north and west of the Isthmus of Tehuantepec (pallidulus and
sumichrasti) versus those to the south and east (costaricensis, decolorus, florencei, nitellinus,
and venustulus). Meanwhile, Corley et al. (2011) found substantial levels of genetic divergence among specimens of Nyctomys, suggesting a possible paraphyletic relationship between taxa and the likely recognition of a new genus.
Firgure 3. Distribution of the genus Nyctomys, with a detailed view in Mesoamerica (obtained from
https://www.gbif.org/)
1.2.3. Genus Scotinomys
Neotropical singing mice of the genus Scotinomys are restricted to the Mesoamerican mountains at elevations above 1000 m. Two species, Scotinomys teguina and S.
xerampelinus, are formally recognized based on the last revision of the genus (Hooper 1972).
According to the literature, Alston's brown mouse (Scotinomys teguina; Alston 1877) is a widespread species inhabiting mid to high-altitude cloud forest, forest edge, and abandoned pastures from southeastern Mexico to western Panama (1000–2900 m). Whereas Chiriquí brown mouse (S. xerampelinus; Bangs 1902) is restricted to the highest forested summits and Páramo of Costa Rica and Panama (2000–3300 m; Wilson & Reeder 2005; Hooper & Carleton
1976). The two ecologically similar congeners have parapatric distributions in the Cordillera de Talamanca and Cordillera Central, where the upper distribution of S. teguina contacts the lower distribution of S. xerampelinus between 2200 and 2900 m (Figure 4; Enders & Pearson 1939; Hooper 1972; Hooper & Carleton 1976). In relation to behaviour and reproduction, both present substantial divergences. Interspecific competition and species differences in thermal tolerance are theorised to maintain this narrow contact zone (Hill & Hooper 1971; Hooper & Carleton 1976).
Figure 4. Distribution of the genus Scotinomys, with a detailed view of Mesoamerica (obtained from
https://www.gbif.org/).
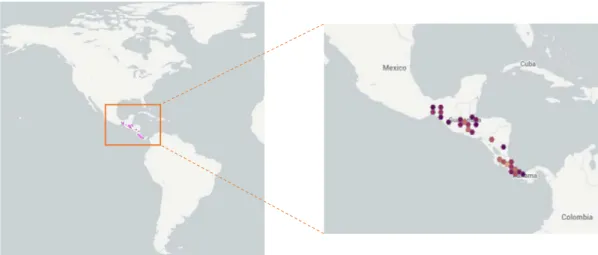
1.2.4. Genus Reithrodontomys
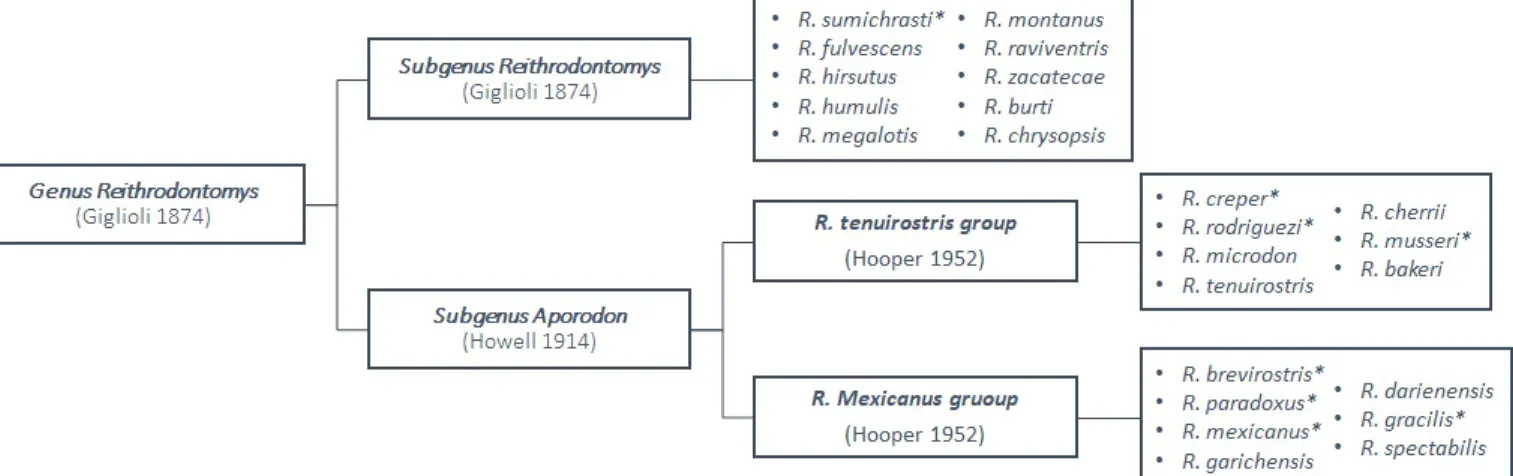
New World harvest mice belonging to the genus Reithrodontomys, are small long-tailed rodents and comprise about 24 species divided in two recognized subgenera (Figure 5):
Reithrodontomys and Aporodon (Hooper 1952; Howell 1914; Arellano et al. 2003). Only after Peromyscus, Reithrodontomys is second in species richness within peromyscines. The
biodiversity of this group reaches its highest point in Mesoamerica, despite of harvest mice being distributed in most habitats in North and Central America as well as in extreme northwestern of South America (Figure 6; Eisenberg 1989; Hall 1981; Hooper 1952). In the genus Reithrodontomys, 12 species are present in southern Mexico alone (Reid 1997): R.
bakeri, R. chrysopsis, R. fulvescens, R. gracilis, R. hirsutus, R. megalotis, R. mexicanus, R. microdon, R. spectabilis, R. sumichrasti, R. tenuirostris, and R. zacatecae. Whereas species
of mesoamerican Reithrodontomys usually have restricted geographic distributions, with the exceptions of R. mexicanus, which occurs from northeastern Mexico south to Ecuador and R.
megalotis, which is found from southwestern Canada to the southern plateau of Mexico. Aside
from R. fulvescens, R. gracilis and R. spectabilis, north and mesoamerican Reithrodontomys typically occupy moderate to high elevation habitats forming “mountain islands” throughout the region. Despite intense interest and rigorous effort, elucidating the relationships within this
FCUP Cryptic diversity in rodents from Costa Rica
9
group has proven to be difficult, and classification within Neotominae has not always been consistent, with the monophyly and composition of lineages remaining unresolved.
Figure 5. Grouping of recognised species of the genus Reithrodontomys per subgenus; * = Species found in Costa
Rica.
Arellano et al. (2003) recently evaluated phylogenetic relationships among some members of the subgenus Aporodon. In addition, Bradley et al. (2004) described a new species of Aporodon (R. bakeri). Arellano et al. (2005) recognised Reithrodontomys cherrii (formerly a subspecies of R. mexicanus) as a deeply divergent clade from the Cordillera de Talamanca, Costa Rica, with affinities to the R. tenuirostris species group. Miller and Engstrom (2008) identified two undescribed species of Reithrodontomys, one from the Cerro de la Carpintera and another from Volcán Poas in Costa Rica. The species-level status of R. cherrii was confirmed by Gardner and Carleton (2009), based on detailed examination of morphological evidence. Hence, the species diversity of Reithrodontomys in southern Central America is greater than what would be assumed from the literature as recent genetic studies have significantly uncovered highly divergent clades that are inconsistent with current understanding of species limits (Sullivan et al. 2000; Arellano et al. 2003; 2005; 2006).
Figure 6. Distribution of the genus Reithrodontomys (obtained from https://www.gbif.org/).
1.2.5. Genus Proechimys
Neotropical spiny rats of the family Echimyidae are considered the most ecologically, morphologically and taxonomically diverse group of all extant South American Hystricognathi rodents. It comprises an old family, with members documented from the Oligocene of Bolivia, approximately 25 million years ago (Patterson & Wood 1982). Among the extant forms, there are about 18 genera and 80 species (Eisenberg & Redford 1999; Emmons & Feer 1997; McKenna & Bell 1997; Woods 1993). They are generally the most abundant and widespread lowland forest small-bodied rats throughout much of their range in neotropical forests (Eisenberg 1989). Within this family, Proechimys is the most diverse genus (Miranda & da Silva 2015), with 25 recognised terrestrial species. The genus ranges from Honduras to Paraguay, and occurs throughout Amazonia (Figure 7), presenting extremely complex systematics, often with reports of four or five species sympatric at a single locality (Patton & Gardner 1972; da Silva 1998). Reconstructing the systematics and phylogeny of Proechimys species have been greatly challenging by extreme levels of intra- and inter-population character variability. Moreover, their interspecific differences in ecological traits and short life cycles allow for a high rate of recovery and diversification of the species in this group, especially in dynamic environments like most tropical ecosystems (Rocha et al. 2014).
Proechimys semispinosus (Central American spiny rat) is the sole member of the genus within
its geographical range in Central America and north-western South America. This species is the best-studied member of the genus and is found in lowland tropical forests ranging from dry to pluvial and from secondary to primary forests (Lambert & Adler 2000).
FCUP Cryptic diversity in rodents from Costa Rica
11
1.3. Molecular markers
Rodents, as the most abundant and diversified order of living mammals, have always been a challenge for researchers attracted by their origins, ways of radiation, and times of diversification (Huchon et al. 2002). Historically, species boundaries and higher taxonomic categorizations within the class Mammalia were based on morphological characteristics (Shoshani & McKenna 1998; Asher 2007), but recent molecular studies led to a better comprehension over the lineages of related species, especially, at lower taxonomic levels, where derived morphological characteristics can be difficult to determine given recent divergences. Therefore, leading to often radically dissimilar phylogenies and species assemblages (Buckley-Beason et al. 2006; Tabuce et al. 2008).
Mitochondrial DNA (mtDNA) possesses a set of unique and valuable characteristics for understanding the evolutionary relationships between individuals, populations, and species (Avise et al. 1987). Among the reasons behind the preference of mtDNA over other genetic markers in animal population studies are its high level of variability, its uniparental (maternal) inheritance, and its practically neutral mode of evolution (Wright 1931). Moreover, the evolutionary rate of the mtDNA is 5 to 10 times faster than the nuclear DNA (nDNA) (Brown et al. 1979), hence being particularly adequate for studying taxa at low taxonomic levels like intra-generic or intra-specific relationships (Hillis et al. 1996). It presents a high level of transitions and transversions, as well as a high incidence of small length mutations (Cann & Wilson 1983) and it is assumed to reflect demographic effects, namely, variations in population size between species or populations, which makes it a popular tool for conservation purposes (Harrison 1989; Roman & Palumbi 2003).
The two most employed mitochondrial markers are Cytochrome b (cytb) and Cytochrome c oxidase I (COI) since these are considered helpful to detect genetic diversity and differentiation at the intra- and inter-specific level between closely related species (Low et al. 2014).
The cytochrome b (cytb) gene has been widely used in many studies for species identification in numerous animal taxa (Irwin et al. 1991; Parson et al. 2000; Hsieh et al. 2001; Rajapaksha et al. 2002; Ono et al. 2007; Tsai et al. 2007; Barbosa et al. 2013). It is probably the best-known mitochondrial gene in studies of phylogenetic relationships for its both slowly and rapidly evolving codon positions, as well as more conservative and more variable regions or domains (Esposti et al. 1993). Therefore, this gene is often used in phylogenetic and phylogeographic studies and several studies of mammalian orders have resulted in new classification structures and have served to assign newly described species to a genus as well
as to enhance the understanding of evolutionary relationships (Sturmbauer & Meyer 1992; Lydeard & Roe 1997; Kumazawa & Nishida 2000; Castresana 2001).
The cytochrome c oxidase subunit I (COI) mitochondrial region has emerged as the standard barcode region for animals proposed by Herbert et al. (2003). The choice of this gene fragment to distinguish known species is based on its high variability, inheritance mode and neutral evolution (Hebert et al. 2003; Ward et al. 2005). It is also widely used to identify new or cryptic species (e.g. Hebert et al. 2003; 2004; Hajibabaei et al. 2006; Smith et al. 2006; Witt et al. 2006), and to assess biodiversity (Janzen et al. 2005; Smith et al. 2005) in many animal groups, including mammals. The efficiency of DNA barcodes relies on the existence of a barcoding “gap” between intra- and interspecific variation. Interspecific divergences in COI sequences are significantly higher than intraspecific variation in many groups of animals, specifically, sequence divergence between most congeneric species is generally greater than 2% (Hebert et al. 2003), whereas intraspecific variation is often lower than 1% (Avise 2000).
Nevertheless, their application is not exempt of problems. The efficiency of using mtDNA in population-genetic studies has been significantly weakened by the presence of mitochondrial pseudogenes (NUMTs), and other factors as incomplete lineage sorting or mitochondrial introgression (Zhang & Hewitt 1996; Bensasson et al. 2001). Therefore, the combination of nuclear (nDNA) and mitochondrial DNA (mtDNA) markers has proven to be more effective to test phylogenetic and phylogeographic hypotheses and emphasized the limitations of studies using mtDNA markers only (Toews & Brelsford 2012).
Moreover, nuclear genes (nDNA) have been also used in vertebrate systematics. Nuclear loci can provide independent estimates of phylogenetic relationships to corroborate or refute phylogenies constructed using mtDNA sequence data (Gaines et al. 2005). Introns are a common source of nuclear sequence data because they evolve with fewer evolutionary constraints relative to coding sequence (Palumbi & Baker 1994; Prychitko & Moore 1997; Friesen et al. 1999; Hare & Palumbi 2003). Exons are also considerably efficient in resolving deep-level mammalian clades (Springer et al. 2001). The Interphotoreceptor Retinoid Binding Protein (IRBP) exon 1 has been used extensively in mammalian phylogeny inference (Springer et al. 1997; Stanhope et al. 1992, 1996), and appears to be useful for discerning relationships at lower taxonomic levels (Jansa & Voss 2000; Weksler et al. 1999). Whereas, the nuclear Recombination Activating Gene 1 (RAG1) has been used for phylogenetic inference at various taxonomic levels (Groth & Barrowclough 1999; Mathee et al. 2004). Several studies have focused on specific vertebrate groups (Hugall et al. 2007; Krenz et al. 2005; Meijden et al. 2004) and have highlighted the characteristics of this gene in relation to its phylogenetic utility. Some of these potentially useful characteristics of RAG1 include its
FCUP Cryptic diversity in rodents from Costa Rica
13
existence as a single copy gene (Evans et al. 2005), uninterrupted exon (Venkatesh et al. 1999). In addition, the conserved nature of certain regions of the gene, especially its second half (3' end), facilitates the design of degenerate "universal" primers for PCR, and the presence of numerous sequences from a variety of taxa in public databases, and an overall lack of saturation (Hugall et al. 2007) promotes its use in phylogenetic studies.
1.4. Objectives
Costa Rica shows an exceptional natural stage for studying the recent historical assembly and diversification of Neotropical rodent biota. This work is integrated in Alexander Gómez Lépiz phD research (“Small mammals distribution and genetic diversity along altitudinal gradients in Costa Rica”) and aims, with the application of molecular approaches, to contribute to the understanding of the genetic diversity and phylogeography of rodent taxa in this area. Namely, this study focuses on some genus of the Cricetidae (Oligoryzomys,
Nyctomys, Scotinomys, Reithrodontomys) and Echimyidae (Proechimys) families, where high
genetic diversity was detected, aiming to provide insights on the phylogenetic relationships and phylogeography of these taxa.
Hence, this study has the following specific goals:
I) Analyse genetic diversity of Cricetidae and Echimyidae rodents in Costa Rica and detect possible events of cryptic speciation;
II) Study the phylogeography of these species in Costa Rica;
Therefore, both mitochondrial (Cytb and COI) and the nuclear (IRBP and RAG1) genetic markers were used to assess the genetic variability as well as to understand the evolutionary history and current status of these species. Furthermore, this will contribute to the DNA barcode reference library of rodents in the region. We expect that the results can be of added value for conservation, as it will allow a better awareness of biodiversity richness in the region, that will be highly relevant for developing conservation strategies for this group.
2. Material and Methods
2.1. Study area and sampling
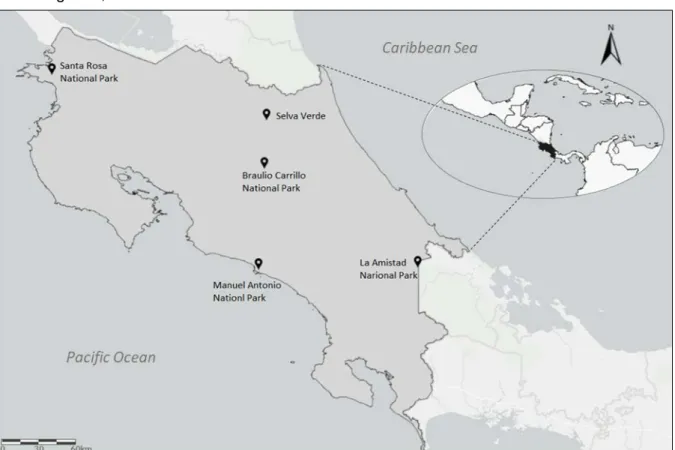
The study area comprises five natural reserves located along Costa Rica (Figure 8), which encompass diverse habitats along altitudinal gradients that consist of rainforests, cloud forests, tropical dry forests and montane surroundings. A total of 624 tissues samples (from museum, ear and tail), in addition to one sample from Honduras, were previously collected within the frame of Alexander Gómez Lépiz PhD work. Due to the large number of samples available, a previous selection of the 221 samples of 11 species, belonging to 5 genera of Cricetidae and Echimyidae families was performed for the analyses of both mitochondrial and nuclear genes, summarised in Table 2.
Figure 8. Map of Costa Rica depicting the collecting localities of the samples of Cricetidae and Echimyidae
available for analyses.
Table 2. Genera, number of species and analysed samples using mitochondrial (cytb and COI) and nuclear (RAG1
and IRBP) genes, with reference to the sampling sites.
Order Family Genus # of
Species
Number of samples
Number of sequences Sampling site
Cytb COI IRBP RAG1
R odent ia Cricetidae Oligoryzomys 2 35 - 15 9 7 LAIP, SRNP Nyctomys 1 9 5 7 4 - LAIP, SV Scotinomys 2 57 29 21 - 4 LAIP, HN, BCNP Reithrodontomys 5 93 64 23 19 18 LAIP, BCNP
Echimyidae Proechimys 1 12 10 6 - 8 LAIP, MANP, SV
(-) No representative samples for analyses; LAIP - La Amistad National Park; SRNP - Santa Rosa National Park; SV - Selva Verde; HN - Honduras; BCNP - Braulio Carrillo National Park; MANP - Manuel Antonio National Park.
FCUP Cryptic diversity in rodents from Costa Rica
15
2.2. DNA extraction, amplification and sequencing
DNA extraction of the selected tissue samples was performed by using EXTRACTME® Genomic DNA 96-Well Kit (DNA GDAŃSK) and eluted twice in 50 µL volume. Whereas DNA from museum samples was extracted following the previously optimized protocol of Dabney et al. (2013). Afterward, quality and quantity of the extracted DNA were evaluated by electrophoresis in TBE (Tris-Borate-EDTA buffer), in 0.8% agarose gel stained with GelRed™ (Biotum), allowing to visualize DNA bands through an ultra-violet (UV) transilluminator (Bio-Rad). The samples with high concentration of DNA were diluted with ultrapure water. Two mitochondrial genes, cytochrome b (cytb) and a fragment of cytochrome c oxidase subunit I (COI), as well as fragments of two nuclear genes, interphotoreceptor retinoid binding protein (IRBP) and recombination activating gene 1 (RAG1) were amplified in the extracted samples (Table 2).
2.2.1. Cytochrome b (cytb)
The cytochrome b (cytb, 1143 bp) fragment was amplified for 108 samples (5
Nyctomys, 29 Scotinomys, 64 Reithrodontomys and 10 Proechimys; see Table 2) using
published primers (Table 3). Protocols were optimized for different taxa mainly adjusting annealing time and temperature. Each performed PCR contained a total volume of 10 µL, composed by 5 µL of Qiagen© PCR Multiplex Kit Master Mix (Qiagen, Hilden, Germany), 0.4 µL of each primer (from a 10 µM solution), 3.2 µL of pure H2O and 1 µL of DNA extraction product (~50 ng/µL). PCR thermal conditions were as follows: initial denaturation for 15 min at 95ºC, followed by 35 cycles of 45 sec at 95ºC for denaturation, 45 sec at the annealing temperature and 1 min at 72ºC, and a final extension for 5 min at 60 ºC. The annealing temperature varied according to the primer pairs used for each genus (Table 4).
Quantity and quality of PCR products were visually examined by 2% gel electrophoresis. Successful amplified PCR products were purified using ExoSAP-IT® PCR clean-up Kit (GE, Healthcare, Piscataway, NJ, USA) to remove excess primers and unincorporated nucleotides during PCR reaction. Sequencing reactions were performed using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA) and subsenquently sequenced for both strands on a 3130XL automated sequencer (Applied Biosystems, USA), or sequenced by GeneWiz Sanger sequencing (USA). All the forward and reverse sequences obtained were aligned and verified manually with software GENEIOUS© v8.1.9 (Bioinformatics Software) using default settings. All sequences were inspected for the presence of stop codons or indels, that could indicate the presence of a potential nuclear mitochondrial
pseudogenes (NUMTs). The sequences suspected to be NUMTs, were not considered for further analyses.
Table 3. Primers used for amplification and sequencing of Cytb gene.
Primer name Primer (5'→3') sequence Direction References
MVZ05M CGAAGCTTGATATGAAAAACCATCGTTG Forward Smith & Patton 1993
MVZ14M TCTTCATCTTTGACTTACAAGG Reverse Smith & Patton 1993
L14724 CGAAGCTTGATATGAAAAACCATCGTTG Forward Irwin et al. 1991
L14727-SP GACAGGAAAAATCATCGTTG Forward Jaarola & Searle 2002
H15915 TCTCCATTTCTGGTTTACAAGAC Reverse Jaarola & Searle 2002
Table 4. Primers pair and annealing temperature optimized in each genus for Cytb gene amplification. Primers pair Genus Annealing temperature
MVZ05M / H15915 Scotinomys 50 ºC Reithrodontomys 50 ºC Proechimys 50ºC L14727-SP / H15915 Nyctomys 52ºC Scotinomys 52ºC Reithrodontomys 52ºC
2.2.2. Cytochrome c oxidase subunit I (COI)
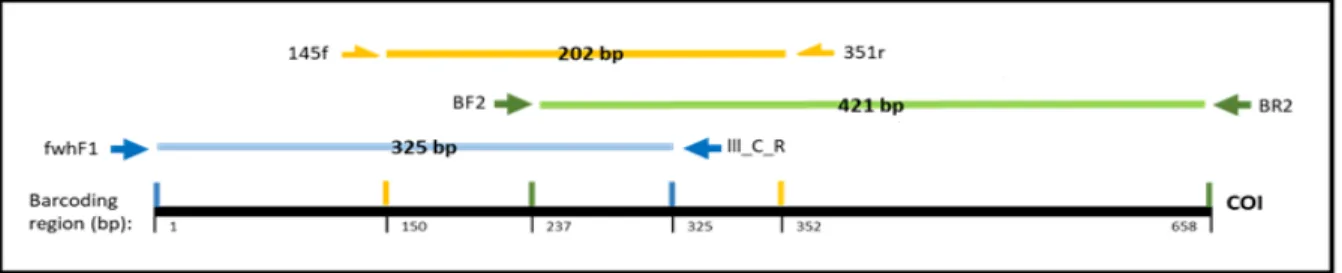
The cytochrome c oxidase subunit I gene (COI, 658 bp) was amplified for 72 samples (15 Oligoryzomys, 7 Nyctomys, 21 Scotinomys, 23 Reithrodontomys and 6 Proechimys; see Table 2), previously selected in order to include all species and sampling sites in the study area. The amplification was performed in two different fragments, using a high throughput sequencing approach, based on a double-indexing pipeline, with two step PCR (Figure 9). The fragments used were: B2 (421 bp) and LCn (325 bp) using published primers sets (Table 5). The first PCR contained a total volume of 10 μL, composed by 5 μL of Qiagen© PCR Multiplex Kit Master Mix (Qiagen, Hilden, Germany), 0.3 μL of each primer (from a 10 μM solution), 3.4 μL of distilled H2O and 1 μL of DNA extraction product (~50 ng/μL). B2 PCR was performed with BF2 / BR2 primer pair (Table 7) with thermal conditions as follows: initial denaturation for 15 min at 95ºC, followed by 40 cycles of 30 sec at 95ºC for denaturation, 45 sec at 51ºC for annealing and 45 sec at 72ºC, and 10 min at 60ºC for final extension. LCn PCR was performed with fwhF1 / Ill_C_R primer pair (Table 7) with thermal conditions as follows: initial denaturation for 15 min at 95ºC, followed by 35 cycles of 30 sec at 95ºC for denaturation, 90 sec at 45ºC for annealing and 45 sec at 72ºC, followed by a final extension of 10 min at 60ºC.
FCUP Cryptic diversity in rodents from Costa Rica
17
Figure 9. COI Primer sets for DNA barcoding of small mammals from Costa Rica. Table 5. Primers used for amplification and sequencing COI fragments B2 and LCn. Primer name Primer (5'→3') sequence Direction References
BF2 GCHCCHGAYATRGCHTTYCC Forward Elbrecht & Leese 2017
BR2 TCDGGRTGNCCRAARAAYCA Reverse Elbrecht & Leese 2017
fwhF1 YTCHACWAAYCAYAARGAYATYGG Forward Vamos et al. 2017
Ill_C_R GGIGGRTAIACIGTTCAICC Reverse Shokralla et al. 2015
For the museum samples, in which DNA was more degraded and failed to amplify B2 or LCn fragments, the SFF primer pair 145f / 351r (Table 6) was used to amplify a smaller fragment of 202 bp. The PCR thermal cycle was performed using the following protocol: initial denaturation for 10 min at 95ºC, followed by 45 cycles of 30 sec at 95ºC for denaturation, 30 sec at 52ºC for annealing and extension of 30 sec at 72ºC, with a final extension of 10 min at 72ºC.
Table 6. Primers used for amplification and sequencing COI fragment SFF. Primer name Primer (5'→3') sequence Direction References
SFF_145f GTHACHGCYCAYGCHTTYGTAATAAT Forward Walker et al. 2016
SFF_351r CTCCWGCRTGDGCWAGRTTTCC Reverse Walker et al. 2016
The quantity and quality of PCR products were visually examined by 2% gel electrophoresis.
After this first PCR, the obtained products were purified using 0.8 μL Beckman Coulter™ Agencourt AMPure XP magnetic beads per 1 μL PCR product, in order to remove primer dimer and unincorporated reagents in the reaction, succeeding two cleaning steps with ethanol, lasting 30 sec each, and a final immersion of purified DNA in 10nM Tris at pH 8.5. Purified products were quantified by using NanoDrop™ 2000 spectrophotometer (Thermo Fisher Scientific) and posteriorly normalized to 10 ng/μL. An indexing PCR was then performed to attach the barcodes in each sample to allow individual identification. PCR reaction was composed by 5 μL of KAPA ReadyMix, 1 μL of compatible Illumina index primers in each pool, 2 μL of H2O and 2 μL of DNA. PCR thermal conditions were: initial denaturation for 3 min at 95ºC, followed by 10 cycles of 30 sec at 95ºC for denaturation, 30 sec at 55ºC for annealing and for extension 30 sec at 72ºC, and a final extension of 5 min at 72ºC. To confirm the success of indexing in all samples, PCR products were visually examined by 2% gel