A diagnosis of
Burkholderia pseudomallei
directly in a bronchoalveolar
lavage by polymerase chain reaction
Manuela Soares Couto
a, Rossana de Aguiar Cordeiro
a,b, Marcos Fábio Gadelha Rocha
b,c,
Thalles Barbosa Grangeiro
d, Natanael Pinheiro Leitão Junior
e,
Tereza de Jesus Pinheiro Gomes Bandeira
f, José Júlio Costa Sidrim
a,b,
Raimunda Sâmia Nogueira Brilhante
a,b,⁎
aPostgraduate Program in Medical Sciences, Federal University of Ceará, Fortaleza, Ceará, Brazil
bSpecialized Medical Mycology Center, Postgraduate Program in Medical Microbiology, Federal University of Ceará, Fortaleza, Ceará, Brazil c
Postgraduate Program in Veterinary Science, State University of Ceará, Fortaleza, Ceará, Brazil
d
Department of Biological Sciences, Federal University of Ceará, Fortaleza, Ceará, Brazil
e
Department of Biological Sciences, State University of Ceará, Fortaleza, Ceará, Brazil
f
LabPauster Laboratory, Fortaleza, Ceará, Brazil
Received 5 February 2009; accepted 11 May 2009
Abstract
Melioidosis is an infectious disease caused by the Gram-negative bacterium Burkholderia pseudomallei. Interest in the molecular identification ofB. pseudomalleihas increased after its classification as a category B agent by the US Centers for Disease Control and Prevention. The present article reports a diagnosis ofB. pseudomalleidirectly in a bronchoalveolar lavage by polymerase chain reaction amplification. The results obtained show that direct detection of the 16-23s spacer sequence in bronchoalveolar lavage is a quick and specific test to diagnose melioidosis.
© 2009 Elsevier Inc. All rights reserved.
Keywords: Melioidosis; Bronchoalveolar lavage; PCR; Diagnosis
Melioidosis is a potentially fatal infection caused by the facultative intracellular bacteriumBurkholderia pseudomal-lei (Merritt et al., 2006). Melioidosis produces diverse clinical manifestations, including acute septicemia, pulmon-ary infection, and chronic visceral and soft tissue abscesses (Supaprom et al., 2007). The clinical symptoms of melioidosis mimic those of many other diseases. Therefore, differentiating between melioidosis and other acute and chronic bacterial infections is often difficult (Peacock et al., 2008). Definitive identification ofB. pseudomalleirelies on an extensive set of biochemical tests that may require up to 5 days before results are obtained.
Practical difficulties for the diagnostic laboratory include the presence of closely related Burkholderia spp.
in specimens from nonsterile sites and atypical colony morphology of some B. pseudomallei strains (Lee et al., 2007). Also, handling of B. pseudomallei cultures is a high-risk activity and is limited to biosafety level 3 laboratories (Peacock et al., 2008). Consequently, the molecular identification may offer a rapid alternative, especially in first-line laboratories equipped to perform DNA amplification methods such as diagnostic polymer-ase chain reaction (PCR) (Merritt et al., 2005). Various PCR procedures for B. pseudomalleihave been developed, but most of them have only been evaluated using pure bacterial cultures (Supaprom et al., 2007). Recently, real-time PCR assays reported the rapid detection of B. pseudomallei through DNA extraction of clinical blood samples (Meumann et al., 2006; Novak et al., 2006; Supaprom et al., 2007). However, although this methodol-ogy is rapid and specific for detection of B. pseudomallei, it is not applicable for routine diagnostic purposes in many laboratories.
Available online at www.sciencedirect.com
Diagnostic Microbiology and Infectious Disease 65 (2009) 73–75
www.elsevier.com/locate/diagmicrobio
⁎Corresponding author. Specialized Medical Mycology Center, Post-graduate Program in Medical Microbiology, Federal University of Ceará, Fortaleza, Ceará CEP: 60.425-540, Brazil. Fax: +55-85-3295-1736.
E-mail address:brilhante@ufc.br(R.S.N. Brilhante).
The present article reports a diagnosis ofB. pseudomallei
directly in a bronchoalveolar lavage by PCR amplification of the specific 16-23s spacer region.
In April 2008, a case of melioidosis in a 17-year-old boy was reported to the Ceará State Health Secretariat. The patient probably contracted the disease in Ubajara (3°51′16″ S, 40°55′16″W), Ceará, in northeastern Brazil, while swimming in a recreational river/waterfall in that region. The bronch-oalveolar lavage obtained from this patient was sent to the Specialized Medical Mycology Center (CEMM) to investi-gate the causative agent of the disease. This was done by PCR assay directly from bronchoalveolar lavage and, concomi-tantly, by culture in blood agar,B. pseudomalleiselective agar, and Ashdown agar. The culture was identified phenotypically according to the criteria described for Gram-negative nonfermentative bacilli and by characteristics such as motility and bipolar stain (Virgínio et al., 2006). The colonies that indicate the possibility ofB. pseudomalleiwere submitted to the oxidase test and colistin (10μg/mL) sensitivity test through
disk diffusion in Müeller–Hinton medium. The bacterium was resistant to colistin and was oxidase positive. Then, the B. pseudomallei diagnosis was confirmed through API20NE (BioMerieux, Marcy I'Etoile, France) (Inglis et al., 2004).
For molecular diagnosis, the total genomic DNA from 2 mL of bronchoalveolar lavage was extracted and purified using a protocol for cell culture from the WIZARD® Genomic DNA purification kit (Promega, Madison, WI), according to the manufacture's instructions. The concentra-tion and purity of the DNA preparaconcentra-tion (170 ng/μL) were
determined by measuring the optical density at 260 and 280 nm using a spectrophotometer (Ultrospec 1100 pro; Amersham Bioscience, Piscataway, NJ). PCR was per-formed according to Merritt et al. (2006) with minor modifications. The reaction was performed in a final volume of 25 μL containing 10 μL of DNA sample (30 ng/μL),
2.5μL of buffer (New England Biolabs, UK), 1 mmol/L of
MgCl2 (Invitrogen, Carlsbad, CA), 50 pmol each of the
primers Bp1 (5′-CGATGATCGTTGGCGCTT-3′) and Bp4 (5′-CGTTGTGCCGTATTCCAAT-3′), 10 mmol/L of pooled deoxynucleoside triphosphates, and 1 U ofTaqpolymerase (New England Biolabs). PCR comprised 4 min at 94 °C, followed by 45 cycles of 30 s at 94 °C, 30 s at 50 °C, and 45 s at 72 °C, with a final hold for 7 min at 72 °C. PCR products were demonstrated by ethidium bromide gel electrophoresis on 1% agar gel and visualized under a ultraviolet light.
A controlB. pseudomalleistrain (CEMM BP 01) used in this study was isolated in northeastern Brazil in 2003 (Rolim et al., 2003) and belongs to the CEMM collection. The controlB. pseudomalleistrain was initially cultured on blood agar medium at 37 °C for 24 to 48 h. A single colony was then inoculated into 10 mL of brain–heart infusion broth and incubated at 37 °C overnight. For the extraction of genomic DNA, the protocol for Gram-negative bacteria of the WIZARD® Genomic DNA purification kit was used, according to the manufacture's instructions. Aliquots of DNA obtained from the control strain and from
bronchoal-veolar lavage after first treatment with buffer lysis were incubated in brain–heart infusion broth at 37 °C for 7 days. The absence of any growth assured the safety of the procedure. Determination of the concentration and purity of the DNA (407.5 ng/μL) and the PCR assay of the control
strain were performed as described for bronchoalveolar lavage specimens.
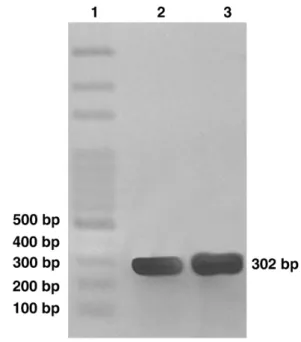
PCR yielded amplification showing products of approxi-mately 300 bp from both bronchoalveolar lavage and the control strain (Fig. 1). Negative control using molecular biology grade water instead of DNA preparations and a bronchoalveolar lavage of 2 patients without suspected for melioidosis did not yield any amplification products.
The present study using PCR assay to detect a specific DNA sequence directly in bronchoalveolar lavage is very interesting because pulmonary melioidosis is the most well-known clinical presentation of the disease (Currie, 2003). Thus, this specific PCR amplification may be a rapid and sensitive method for diagnosis of melioidosis.
The PCR amplification of DNA from both the bronch-oalveolar lavage and control strain showed a single am-plification product of 302 bp.Merritt et al. (2006)identified correctly 71 of 72B. pseudomalleiisolates by PCR analysis, which is able to differentiate B. pseudomallei from other bacteria, including Burkholderia cepacia, Burkholderia multivorans,Burkholderia thailandensis,Burkholderia viet-namiensis, Pseudomonas aeruginosa, Escherichia coli,
Staphylococcus aureus, Staphylococcus epidermidis, and
Mycobacterium tuberculosis.
DNA–DNA homology and cellular lipid and fatty acid composition revealed that B. pseudomallei and
Burkhol-Fig. 1. Agarose gel electrophoresis of PCR products produced from
B. pseudomallei. Lane 1, 100-bp ladder marker; 2, bronchoalveolar lavage sample; 3, control culture (strain number CEMM BP 01). The 302-bp PCR amplification product specific forB. pseudomalleiis indicated.
deria malleiare very closely related to each other. The high homology between the 2 species in DNA sequences is shown in genes, such as flagellin and 16S ribosomal RNA, thus, making it difficult to differentiate them by molecular identification systems (Lee et al., 2005).
AlthoughB. pseudomalleiandB. malleiare very closely related, the importance of this PCR protocol is not impaired, because they differ considerably in other aspects, including ecologic behavior. For instance,B. pseudomalleiis to a large extent a free-living organism, readily isolated from soil and surface water in endemic areas and only on rare occasions finds a niche in animal hosts.B. mallei, however, behaves more successfully with little host specificity and is considered to be an obligate parasite by some investigators. One main morphologic distinction between these 2 species is the presence of flagella inB. pseudomalleiand their absence inB. mallei(Anuntagool and Stitaya, 2002).
Melioidosis has been considered an emerging disease in Brazil since the first cases reported in the northeast region (Rolim et al., 2005). Thus, the recent detection of melioidosis in northeastern Brazil highlights the extent of its distribution in the Americas and underlines the need for improved diagnostic methods (Inglis et al., 2006). Currently, the greater reliability, shorter time for results, simplicity, and lower cost of conventional PCR make it the leading method to confirmB. pseudomalleidiagnosis.
The present study used an alternative method for safe DNA extraction from bronchoalveolar lavage, and this protocol may reduce the time required to obtain definitive diagnostic results, respecting the presumptive diagnosis of melioidosis, which correctly identifiedB. pseudomalleieven before the confirmation by culturing, which occurred after 5 days. Also, it is easily applicable for routine diagnostic purposes in many laboratories in developing countries, which have our reality.
Acknowledgments
This work was supported by the Brazilian National Research Council (CNPq) (grant 475683/2006-4) and FUNCAP (9836/06—Conv. 006-00/2006).
References
Anuntagool N, Stitaya S (2002) Antigenic relatedness between Burkhol-deria pseudomallei and Burkholderia mallei. Microbiol Immunol
46:143–150.
Currie BJ (2003) Melioidosis: an important cause of pneumonia in residents of and travellers returned from endemic regions. Eur Respir J
22:542–550.
Inglis TJJ, Foster NF, Gal DPK, Mayo MJ, Norton R, Currie BJ (2004) Preliminary report on the northern Australian melioidosis environmental surveillance project.Epidemiol Infect132:813–820.
Inglis TJJ, Rolim DB, Sousa AQ (2006) Melioidosis in the Americas.Am J Trop Med Hyg75:947–954.
Lee M, Wang D, Yap EH (2005) Detection and differentiation of Burkhol-deria pseudomallei,Burkholderia malleiandBurkholderia thailanden-sisby multiplex PCR.FEMS Immunol Med Microbiol43:413–417. Lee S, Chong C, Lim B, Chai S, Sam K, Mohamed R, Nathan S (2007)
Burkholderia pseudomalleianimal and human isolates from Malaysia exhibit different phenotypic characteristics.Diagn Microbiol Infect Dis
58:263–270.
Merritt A, Inglis TJJ, Chidlow G, Aravena-Roman M, Harnett G (2005) Comparison of diagnostic laboratory methods for identification of Bur-kholderia pseudomallei.J Clin Microbiol43:2201–2206.
Merritt A, Inglis TJJ, Chidlow G, Harnett G (2006) PCR-based identification of Burkholderia pseudomallei. Rev Inst Med Trop S Paulo48:239–244.
Meumann EM, Novak RT, Gal D, Kaestli ME, Mayo MJ, Hanson JP, Spencer E, Glass MB, Gee JE, Wilkins PP, Currie BJ (2006) Clinical evaluation of a type III secretion system real-time PCR assay for diagnosing melioidosis.J Clin Microbiol44:3028–3030.
Novak RT, Glass MB, Gee JE, Gal D, Mayo MJ, Currie BJ, Wilkins PP (2006) Development and evaluation of a real-time PCR assay targeting Bart J the type III secretion system ofBurkholderia pseudomallei.J Clin Microbiol44:85–90.
Peacock SJ, Schweizer HP, Dance DAB, Smith TL, Gee JE, Wuthiekanum V, DeShazer D, Steinmetz I, Tan P, Currie BJ (2008) Management of accidental laboratory exposure to Burkholderia pseudomalleiand B. mallei.Emerg Infect Dis14:e2.
Rolim DB, Vilar DCFL, Sousa AQ, Miralles IS, Oliveira DCA, Harnett G, O'Reilly L, Howard K, Sampson I, Inglis TJJ (2005) Melioidosis, northeastern Brazil.Emerg Infect Dis11:1458–1460.
Supaprom C, Wang D, Leelayuwat C, Thaewpia W, Susaengrat W, Koh V, Ooi EE, Lertmemongkolchai G, Liu Y (2007) Development of real-time PCR assays and evaluation of their potential use for rapid detection of
Burkholderia pseudomalleiin clinical blood specimens.J Clin Microbiol
45:2894–2901.
Virgínio CG, Teixeira MFS, Frota CC, Café VS, Rocha MFG, Sidrim JJC (2006) Phenotypic characterization of three clinical isolates of Bur-kholderia pseudomallei in Ceará, Brazil. Mem Inst Oswaldo Cruz
101:95–97.
75