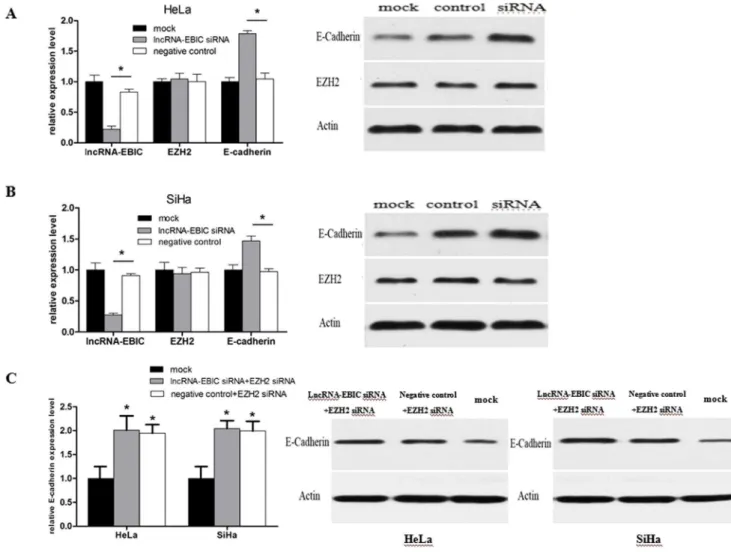
Long noncoding RNA-EBIC promotes tumor cell invasion by binding to EZH2 and repressing E-cadherin in cervical cancer.
Texto
Imagem


Documentos relacionados
The expression of (A) known long protein-coding (long NM_ RefSeq genes) and (B) known lncRNA genes (long NR_ RefSeq genes) and (C) putative embryonic brain lncRNAs over ES cell
Differential expression of 4 of 6 genes found to be differentially expressed by cDNA microarray analysis in the CSIII tumor versus normal tissue com- parison was validated
Using this cell model, we performed microarray assays to compare the miRNA and mRNA expression profiles of Hep-2 and Hep-2/v cells and identified seven differentially expressed
The objective of this study was to evaluate E-cadherin, Slug and neural cell adhesion molecule (NCAM) expression in GH-secreting pituitary adenomas and its relationship to
Assim sendo, acredita-se que os clientes da marca francesa continuam a comprar no seu website por estarem comprometidos com esta, o que significa que a lealdade dos clientes da
Assim, em jeito de conclusão podemos dizer que apesar das muitas debilidades apresentadas e a falta de estruturação do local, este pode e tem condições para se tornar um ponto
jejuni strains isolated from non-human primates during animal management in the Cecal/Fiocruz, RJ, was able to adhere to or invade HeLa cells.. The difference
RNA-Seq analysis and identification of differentially expressed genes (DEGs) - We performed RNA-Seq anal- yses to evaluate spatiotemporal gene expression profiles along