Identification of Gene Modules Associated with Low Temperatures Response in Bambara Groundnut by Network-Based Analysis.
Texto
Imagem

Documentos relacionados
The integration of functional gene annotations in the analysis confirms the detected differences in gene expression across tissues and confirms the expression of porcine genes being
Our microarray analysis of the gene expression pro- file of degenerated IVD detected hundreds of differentially expressed genes that may be associated with IVD degener-
The storage susceptibility of Bambara groundnut (B. G.) ( Voandzeia Subterranean (L.) Thouars) to Callosobruchus maculatus and chemical and functional properties of 11 varieties
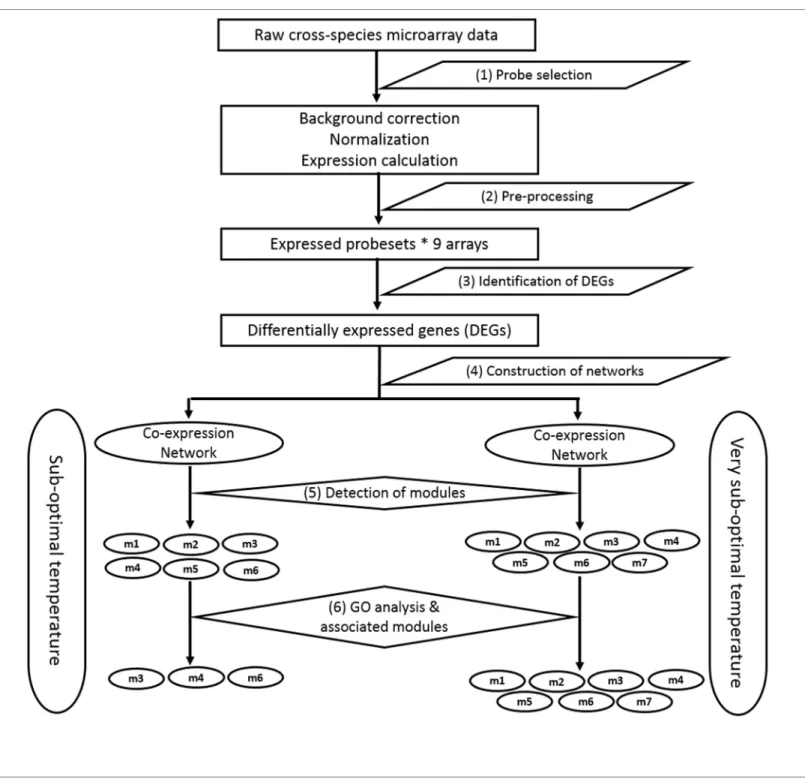
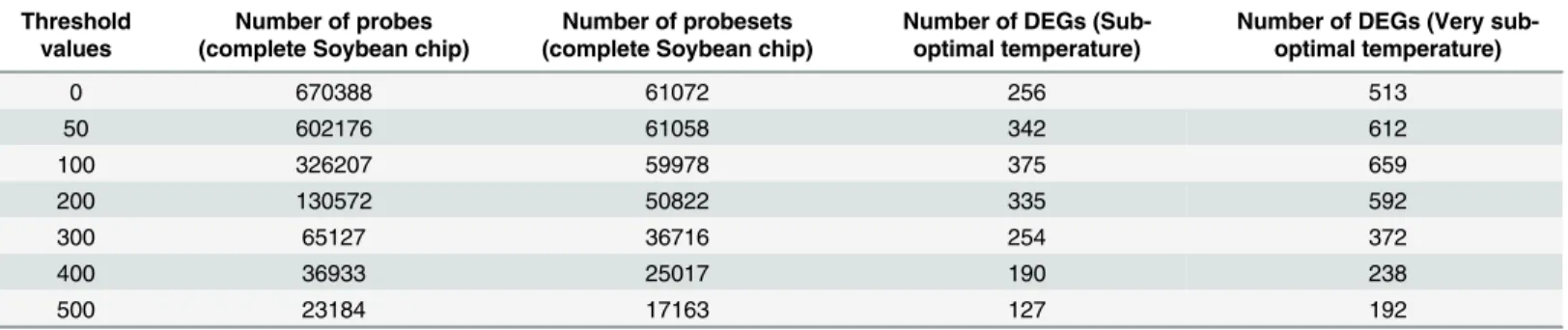
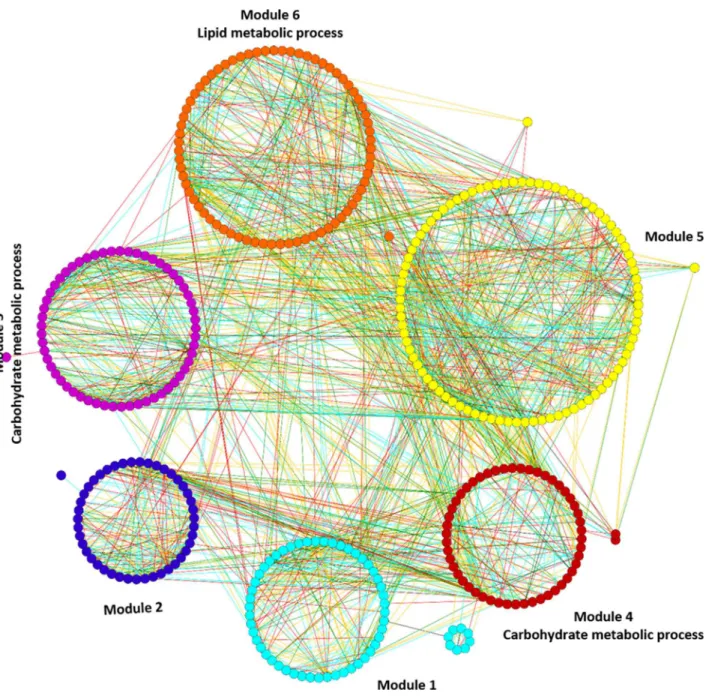
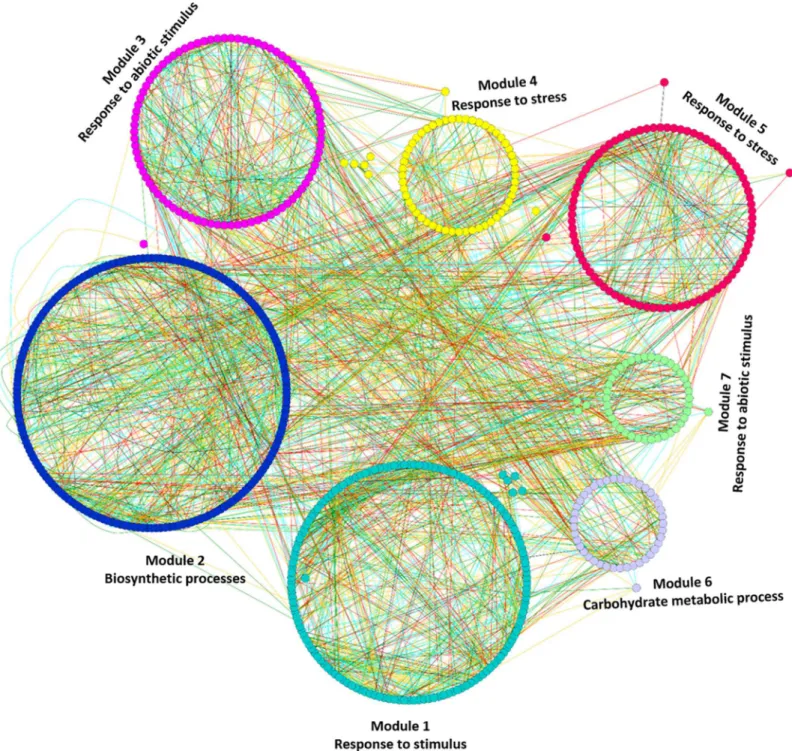
Results: Microarray based gene expression data and protein-protein interaction (PPI) databases were combined to construct the PPI networks of differentially expressed (DE) genes in
Functional identification of gene networks was performed using Ingenuity Pathway Analysis program as previously described [39].The tables representing the differentially
The effects of NaCl salinity on seed germination, growth, physiology, and biochemistry of two bambara groundnut landraces ( Vigna subterranea (L.) Verdc), Kakamega (white seed
Comparative genome-wide expression profiling of malignant tumor counterparts across the human-mouse species barrier has a successful track record as a gene discovery tool in
RNA-Seq analysis and identification of differentially expressed genes (DEGs) - We performed RNA-Seq anal- yses to evaluate spatiotemporal gene expression profiles along
![Fig 4. Co-expression relationships of MYB173 and GATA9 transcription factors with their related genes: [A] Shows the co-expression relationships of MYB173 with its related genes](https://thumb-eu.123doks.com/thumbv2/123dok_br/16352753.189679/13.918.62.864.113.462/expression-relationships-transcription-factors-related-expression-relationships-related.webp)