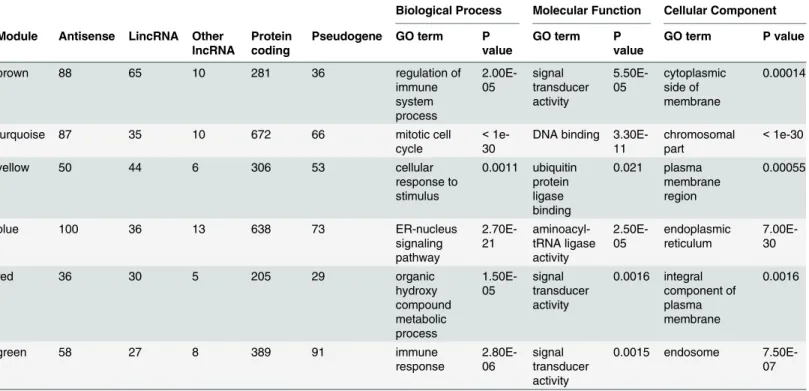
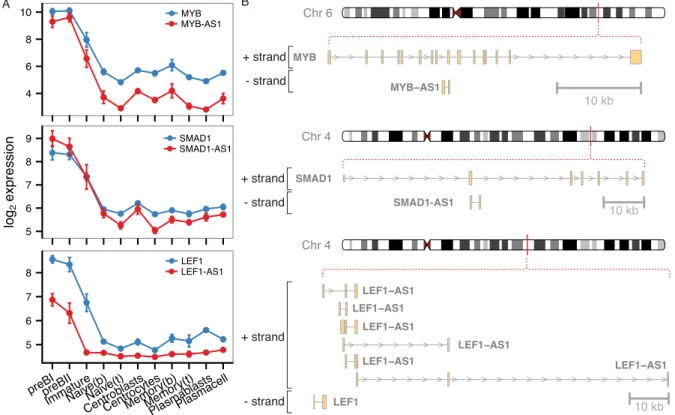
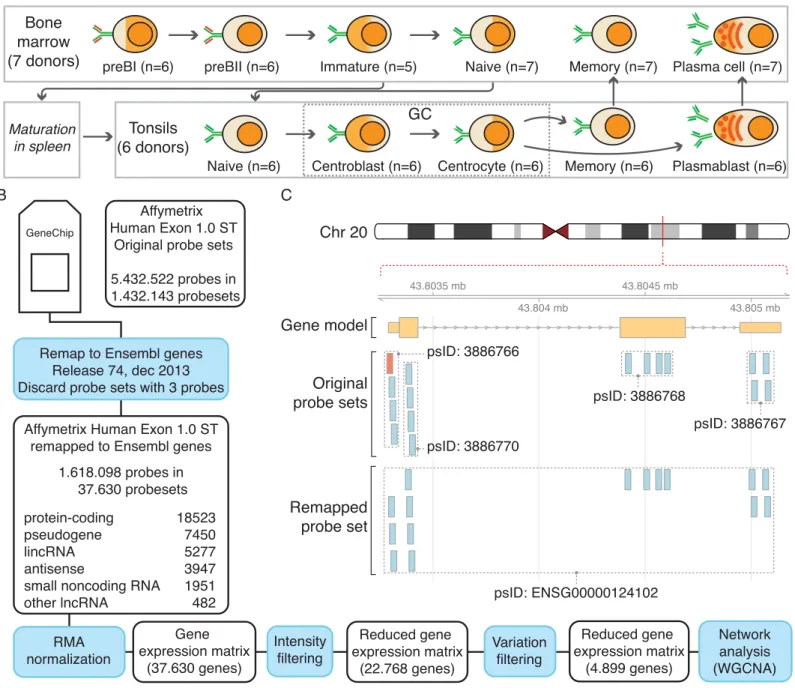
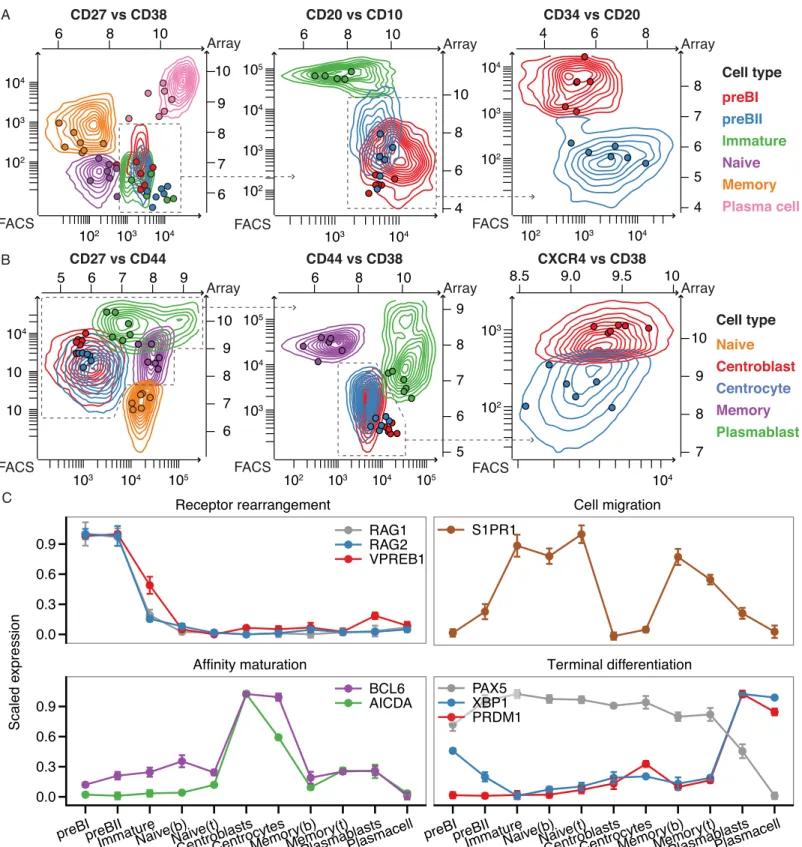
Long Noncoding RNA Expression during Human B-Cell Development.
Texto
Imagem
Documentos relacionados
However, the RNA-seq transcript profiling experiments the differences in expression of a gene between two samples are in fact differences in expression per unit of RNA or “ per
Here, we investigated the protein-coding capacities and the expression levels of their isoforms for human known genes, the conservation and disease association of long noncoding
Figure 7. Dlk1 and myostatin display an inverse relationship in human myoblasts, and the expression of Dlk1 is observed in human rhabdomyosarcoma cell lines. A: Expression of Dlk1
In PBS-treated mice, 1,973 genes (14.8% of 13,326 expressed genes) were significantly upregulated, and 1,915 genes (14.3%) were significantly downregulated at 48 hours after
Here, in order to explore the potential lncRNAs involved in hepatocellular carcinoma (HCC) oncogenesis, we performed lncRNA gene expression profile analysis in 3 pairs of human HCC
The aim of this study was to examine HOTAIR expression in patients with esophageal squamous cell cancer (ESCC) and explore its clinical significance.. Methods: Differences in
To profile differentially expressed lncRNAs in hepatoblastoma, we performed a genome-wide analysis of lncRNA and mRNA expression in hepatoblastoma and matched normal tissues
controlled by PABPN1 at the level of 3 9 end processing [16,45] and the genes identified in our study that showed increased and decreased mRNA levels in PABPN1–depleted cells.