U
NIVERSIDADE DE
L
ISBOA
F
ACULDADE DE
C
IÊNCIAS
D
EPARTAMENTO DE
B
IOLOGIA
V
EGETAL
Screening of antibiotic resistance
determinants in Gram-negative bacteria
isolated from environmental reservoirs
SOFIA RÊGO DE CARVALHO
M
ESTRADO EM
M
ICROBIOLOGIA
A
PLICADA
2011
U
NIVERSIDADE DE
L
ISBOA
F
ACULDADE DE
C
IÊNCIAS
D
EPARTAMENTO DE
B
IOLOGIA
V
EGETAL
Screening of antibiotic resistance
determinants in Gram-negative bacteria
isolated from environmental reservoirs
Dissertação orientada por Dr. Patrick Freire (LNIV-INRB)
e Prof. Dr.ª Lélia Chambel (FCUL)
SOFIA RÊGO DE CARVALHO
M
ESTRADO EM
M
ICROBIOLOGIA
A
PLICADA
2011
Screening of antibiotic resistance
determinants in Gram-negative bacteria
isolated from environmental reservoirs
Sofia Rêgo de Carvalho
M
ESTRADO EM
M
ICROBIOLOGIA
A
PLICADA
2011
Esta tese foi desenvolvida no Laboratório Nacional de Investigação
Veterinária sob a orientação do Dr. Patrick Freire.
A Prof. Drª Lélia Chambel foi a orientadora interna no âmbito do
Mestrado em Microbiologia Aplicada da Faculdade de Ciências da
Universidade de Lisboa.
Agradecimentos
Quero agradecer a todas as pessoas que directa ou indirectamente contribuíram para o meu trabalho desenvolvido no âmbito desta tese de mestrado. Antes de mais quero agradecer ao Dr. Nuno Canada, Director do LNIV-INRB, por me ter aceite nesta instituição.
Em primeiro lugar agradeço ao Dr. Patrick Freire por me ter orientado ao longo deste último ano, por me ter ajudado sempre que eu precisei e por ter tornado a realização desta tese possível apesar dos contratempos.
Gostaria também de agradecer à Prof. Lélia Chambel por ter aceite ser a minha orientadora interna, por se ter mostrado sempre disponível para esclarecer as minhas dúvidas e ajudar na correcção desta dissertação.
Não posso deixar de agradecer ao Dr. Gonçalo da Costa do Centro de Química e Bioquímica da FCUL pela colaboração na realização dos estudos de proteómica.
Aos meus colegas do LNIV pelo excelente ambiente de trabalho e boa disposição que sempre criaram, sem vocês as horas de almoço teriam certamente sido mais curtas. A Inês Guinote merece um agradecimento em especial por toda ajuda que me deu e por dar música ao nosso gabinete
À Inês Albuquerque, Patrícia Almeida, Margarida Bárbaro, Andreia Nunes, Joana Silva e Raquel Jacinto a FCUL contribuiu para minha formação académica mas também me permitiu conhecer um grupo de amigas fantástica.
Gostaria de agradecer aos meus amigos Sandra, Isa, João e João por todos os momentos de diversão e descontracção fora da vida académica.
O Bernardo merece um agradecimento muito especial, não só por me ouvir nos meus momentos microbióloga mas sobretudo por ter estado lá para mim sempre que eu precisei. Obrigada és um espectáculo.
O maior obrigado de todos vai para os meus pais por sempre me apoiarem e proporcionarem tudo, pela infinita paciência e compreensão para comigo. Quero também agradecer ao meu irmão, avôs, tios e restantes membros da família.
Most used Abbreviations
ATCC: American Type Culture Collection bp: base pairscfu: colony forming unit
CLSI: Clinical and Laboratory Standard Institute ESBL: extended-spectrum β-lactamase
EUCAST: The European Committee on Antimicrobial Susceptibility Testing exp: exponential growth phase
HGT: horizontal gene transfer LB: Luria-Bertani
MDR: multidrug resistance MH: Mueller-Hinton
MIC: minimum inhibitory concentration OD: optical density
PMQR: Plasmid Mediated Quinolone Resistance QRDR: Quinolone Resistance Determinant Region rpm: rotations per minute
SDS: sodium dodecyl sulfate
SDS-PAGE: sodium dodecyl sulfate polyacrylamid gel electrophoresis stat: stationary growth phase
1D gel: one dimensional gel-based electrophoresis 2D gel: two dimensional gel-based electrophoresis
½ MIC exp: cells exposed to half of their ciprofloxacin MIC in the exponential growth phase ½ MIC stat: cells exposed half of their ciprofloxacin MIC in the stationary growth phase MIC exp: cells exposed to their ciprofloxacin MIC in the exponential growth phase MIC stat: cells exposed to their ciprofloxacin MIC in the stationary growth phase
2xMIC exp: cells exposed to twice their ciprofloxacin MIC in the exponential growth phase 2xMIC stat: cells exposed to twice their ciprofloxacin MIC in the stationary growth phase
ABSTRACT
Antibiotics are one of the most successful forms of chemotherapy and saved million of lives placing most bacterial infectious diseases under control. However, this success has been compromised the continuous selective pressure exerted by antibiotics use has resulted in multi-resistance bacteria bearing resistance mechanisms to several antibiotics.
Nowadays, there is an increased recognition that not only clinical settings constitute resistance reservoirs. The soil is a vast reservoir of resistance mechanisms and their associated genes, so understanding the resistant determinants present in the soil, the soil resistome, will provide information about antibiotic resistance frequencies and also emergence of new resistance mechanisms.
Quinolones are an important group of synthetic antibiotics, recently plasmid mediated quinolone resistance mechanisms were established, some originated in environmental reservoirs.
In this work the main goal was to study a collection of (fluoro)quinolone resistant environmental isolates, in terms of their ciprofloxacin MIC and screen them for the presence of qnr genes. With a smaller group of isolates the phenotypic impact of ciprofloxacin in growth curves/cellular viability was studied and finally detected and characterized induced variations of cell protein profiles, to study how ciprofloxacin affects, at a molecular level these bacteria.
Results showed that ciprofloxacin MIC values had similar distributions in the soil samples from the two sample types from where these strains were isolated; in terms of resistance determinants results point that a qnrS gene was found in a Comamonas testosteroni isolate. In the second part of this work, Stenotrophomonas maltophilia response to ciprofloxacin was studied, and the growth curves/cellular viabilities results showed that S. maltophilia cells tolerate/resist ciprofloxacin action when in stationary growth. Significant differences were observed between cell total protein profiles from cells exposed to ciprofloxacin and the control. That study can help understand bacterial general response to fluoroquinolones, in addition to more classical genetic mutation, and also what intracellular pathways this antibiotic might trigger.
Keywords: Environmental isolates, antibiotic resistance, ciprofloxacin, S. maltohpilia, protein
RESUMO
A utilização de antibióticos permitiu salvar milhões de vidas e controlar inúmeras doenças infecciosas, contudo a eficácia destes fármacos está hoje em risco. A utilização generalizada dos antibióticos levou ao aparecimento e disseminação de diversos mecanismos de resistência aos antibióticos que comprometem a eficácia dos mesmos. As bactérias multi-resistentes estão a tornar-se um grave problema de saúde pública, pois possuem mecanismos de resistência para várias classes de antibióticos, restando assim poucos antibióticos eficazes para a sua terapêutica.
Durante as últimas décadas, o estudo da resistência aos antibióticos focou-se sobretudo nos meios clínicos e em microrganismos patogénicos. Mas, recentemente, tem sido reconhecido que outros ambientes e microrganismos não patogénicos também contribuem largamente para o processo de desenvolvimento e disseminação de mecanismos de resistência. Um dos ambientes cuja importância em termos de reservatório de resistência tem sido reconhecida é o solo. É um ecossistema complexo onde os antibióticos e os seus genes de resistência sempre estiveram presentes devido aos microrganismos produtores de antibióticos. Contudo, diversas actividades antropogénicas provocam a contaminação do solo com antibióticos e microrganismos resistentes o que influencia não só a estrutura da microbiota do solo como contribui para o desenvolvimento de mecanismos de resistência aos antibióticos. Num estudo recente, em que se analisou o nível de resistência de bactérias do solo a diversas classes de antibióticos, verificou-se que a maioria delas é multi-resistente e são resistentes mesmo a antibióticos sintéticos como as quinolonas. Este e outros estudos contribuíram para o reconhecimento da importância do solo como um reservatório para a emergência de mecanismos de resistência e bactérias resistentes. Surgiu assim um novo conceito, o resistoma do solo que consiste no conjunto de determinantes de resistência presentes no solo.
As quinolonas são um grupo de antibióticos sintéticos que inibe a replicação e transcrição do DNA bacteriano conduzindo à morte celular. Durante muitos anos, pensou-se que a resistência às quinolonas se devia apenas a mutações que modificassem as enzimas alvo (topoisomerases tipo II) ou activassem a expressão de bombas de efluxo. Contudo, nos últimos anos novos mecanismos de resistências às quinolonas codificados em plasmídeos têm sido identificados. Um desses mecanismos é o gene qnr, codifica uma proteína que protege o DNA e as topisomerases tipo II do efeito das quinolonas.
Recentemente a proteómica tem sido utilizada como uma nova abordagem no estudo da resposta bacteriana a diversos estímulos do meio, entre os quais podemos considerar a exposição aos antibióticos. O proteoma é o conjunto de proteínas produzidas por uma
bactéria num determinado momento; ao contrário do genoma, o proteoma é dinâmico e varia rapidamente em função dos diferentes estímulos do ambiente onde as bactérias se encontram. A comparação entre perfis proteicos de bactérias não expostas e expostas a antibióticos está a ser utilizada no estudo dos mecanismos de resistência e resposta das bactérias aos antibióticos ou no modo de acção dos mesmos.
O objectivo geral deste trabalho foi estudar uma colecção de bactérias ambientais resistentes às (fluoro)quinolonas, isoladas de solos provenientes de dois tipos de locais, quintas de produção animal e margens do rio Tejo, onde diversos factores podem contribuir para o desenvolvimento de resistências. Primeiro, esta colecção de isolados foi caracterizada em termos da sua resistência à ciprofloxacina e foi feita uma triagem do gene qnr. Para a execução do segundo objectivo deste trabalho realizaram-se curvas de crescimento e viabilidades para estudar de que forma a ciprofloxacina afecta o crescimento de Stenotrophomonas maltophilia. Perfis protéicos totais de culturas de S. maltophilia crescidas sem ou com ciprofloxacina foram comparados para determinar variações que possam reflectir as mudanças fisiológicas desencadeadas por este antibiótico.
O nível de resistência dos isolados foi estabelecido pela determinação da sua CMI (concentração mínima inibitória) para a ciprofloxacina. Comparando os valores de CMI obtidos entre os isolados provenientes dos dois tipos de locais de amostragem, margens do rio Tejo e quintas de produção de animais, verificamos que são semelhantes, sendo que a maioria das CMI é entre 1 e 8 μg/ml. Para pesquisar a presença de isolados qnr-positivos nesta colecção de bactérias ambientais efectuamos um PCR multiplex com três conjuntos de primers previamente descritos, desenhados para amplificar as principais famílias do gene qnr estudadas até agora: qnrA, qnrB e qnrS. Dos 112 isolados só para um, mais tarde identificado como Comamonas testosteroni, se obteve amplificação positiva. Contudo estes resultados apresentam alguma reserva pois a amplificação não foi totalmente específica e só estudos adicionais permitirão confirmar se este se trata ou não do primeiro gene qnr encontrado em C. testosteroni.
Para a realização do segundo objectivo principal deste trabalho o grupo de estirpes em estudo foi reduzido. Primeiro, os isolados com maior CMI para a ciprofloxacina foram identificados por galerias API®. Estes 16 isolados pertencem a 4 géneros diferentes,
Comamonas, Acinetobacter, Stenotrophomonas e Aeromonas. Seguidamente para os isolados seleccionados foram determinadas as CMI e os halos de inibição para alguns antibióticos da classe dos β-lactâmicos. Os resultados revelaram que os isolados S. maltophilia são os que apresentam maior nível de resistência a esta classe de antibióticos.
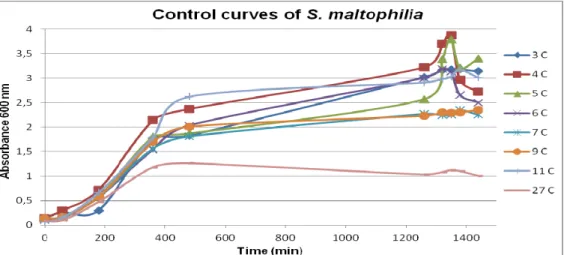
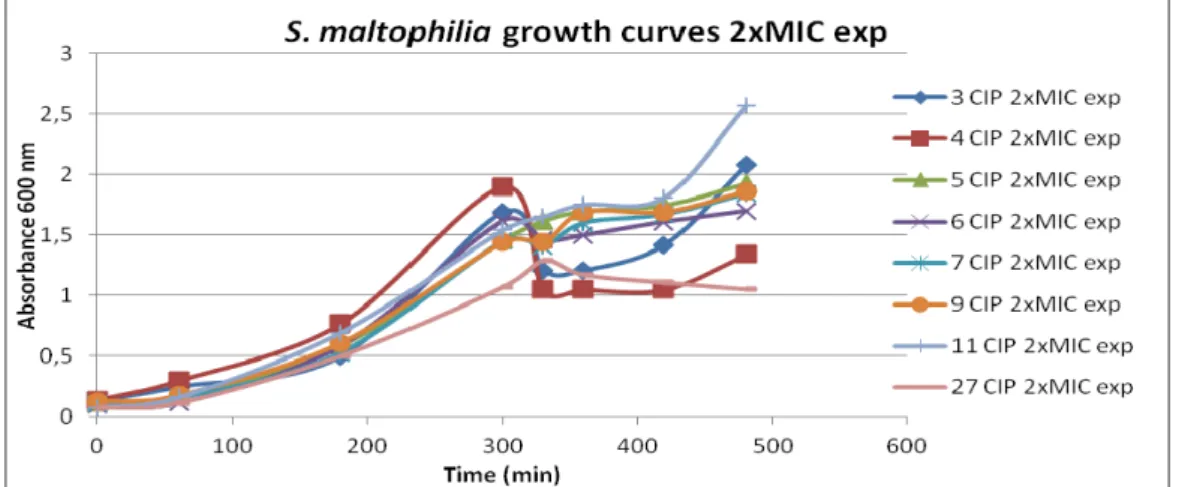
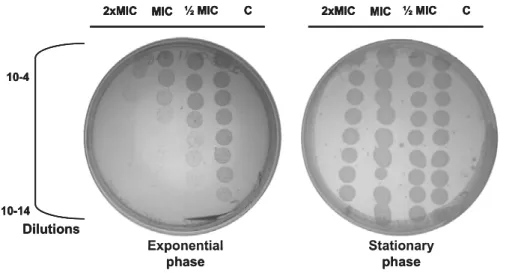
Com base em todos os resultados obtidos previamente optou-se por utilizar os oito isolados de S. maltophilia para a realização do segundo objectivo principal deste trabalho. Primeiro foram realizadas curvas de crescimento em que diferentes quantidades de ciprofloxacina, correspondentes a 1/2MIC, MIC e 2xMIC de cada isolado, eram adicionadas a culturas de S. maltophilia durante a fase exponencial de crescimento ou durante a fase estacionária de crescimento. Em paralelo, diluições da cultura foram plaqueadas para testar como a ciprofloxacina estava a afectar a viabilidade celular. Em conjunto estes resultados permitiram verificar que as curvas de crescimento por OD não reflectem o efeito que a ciprofloxacina está a ter nas células S. maltophilia. Quando a ciprofloxacina é adicionada, durante a fase exponencial, a viabilidade celular decresce significativamente comparativamente ao controlo mas tal descida não se reflecte numa descida de OD600nm nas
curvas de crescimento. A análise dos resultados obtidos quando a ciprofloxacina é adicionada a bactérias que já se encontram em fase estacionária indica que nesta fase estas toleram e/ou resistem à presença de ciprofloxacina pois o tratamento com este antibiótico não provoca morte celular.
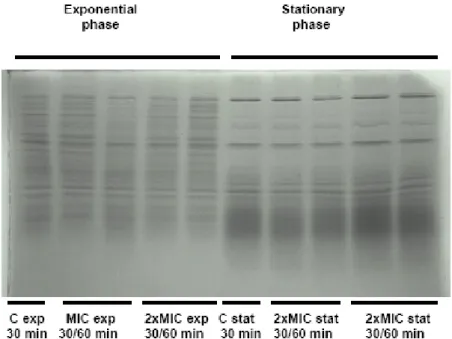
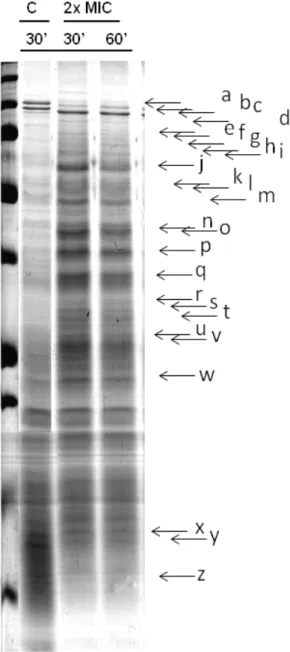
Perfis proteicos de culturas de S. maltophilia crescidas em diferentes condições foram obtidos por SDS-PAGE. Primeiro num sistema de mini géis com o qual os géis obtidos não permitiram uma suficiente discriminação do padrão mas revelaram uma clara distinção entre os perfis de células em fase exponencial ou estacionária. Visto as células de S.maltophilia serem afectadas pela ciprofloxacina, quando em crescimento exponencial, os perfis proteicos das células nestas condições foram repetidos em géis de maiores dimensões. Deste modo, foi possível obter perfis proteicos com maior resolução e comparar os perfis proteicos das células em condições controlo com as expostas à ciprofloxacina. Esta comparação revelou diferenças significativas, especialmente para o caso do isolado S. maltophilia 4 indicando que os perfis proteicos reflectem o ambiente de crescimento das bactérias e nomeadamente o stress induzido pela ciprofloxacina. O estudo mais pormenorizado destas variações do proteoma, recorrendo a técnicas como géis 2D e espectofotometria de massa, está em curso e permitirá identificar proteínas com potencial relevância nos processos de resistência ou no modo de acção da ciprofloxacina.
Palavras-chave: Isolados ambientais, resistência aos antibióticos, ciprofloxacina,
CONTENTS
Agradecimentos ...I Most used Abreviations ...II Abstract...III Resumo ... IV Contents... VII
Introduction ...1
Mechanisms of antibiotic resistance ...2
Evaluation of microorganim’s susceptibility testing ...3
Antibiotic resistance in the environment ...3
Presence of antibiotics in environmental settings ...4
Quinolones / Fluoroquinolones ...5
Quinolones resistance mechanisms ...7
β-Lactams resistance ...9
Proteomics and antibiotic resistance ...10
Aims...12
Materials and Methods...13
Bacterial strains ...13
Antibiotic susceptibility studies ...13
Identification of isolated strains ...14
S. maltophilia and ciprofloxacin ...14
Growth curves and celular viabilities...14
Protein profiles ...15
Total DNA extraction ...15
Screening for Qnr-positive isolates...16
Results and Discussion...17
Environmental isolates resistant to (fluoro)quinolones ...17
Determination of ciprofloxacin MIC ...17
Screening for Qnr-positive isolates ...19
Isolates selection and identification ...19
Resistance to β-Lactams...21
S. maltophilia and ciprofloxacin ...24
Growth curves and celular viabilities...24
Protein profiles ...28
Conclusion and Future Prespectives ...32
Annex ...34
INTRODUCTION
Antibiotics are probably one of the most successful forms of chemotherapy in the history of medicine. Millions of lives have been saved and the majority of bacterial infectious diseases placed under control 1. An antibiotic is a substance of biological, semi-synthetic or synthetic
origin, which shows selective activity against bacteria and thus can be used in the treatment of infectious diseases 2. Antibiotics can be classified based on the cellular component or
system they affect. They are defined as bactericidal when antibiotic exposure leads to cell death, or bacteriostatic if antibiotic exposure inhibits growth, without viability loss 3. Three
preferential mechanisms were established as targets for the major antibiotic groups: cell-wall biosynthesis, RNA and protein machinery and DNA synthesis and repair 3,4.
When introduced into clinical practice in the 1940´s, antibiotics were extremely efficient in treating bacterial infectious 1. However, antibiotic’s success has been compromised. Due to
spreading of antibiotic resistance mechanisms among pathogens, antibiotics are now ineffective against an increased number of bacterial infections. New antibiotics enter into regular therapeutic used after years of clinical trials and safety requirements; clinically significant resistance appears in periods from months to few years. Continued selective pressure exerted by the use of different antibiotics has resulted in microorganisms bearing resistance mechanisms to one or several drugs, leading to the problem of multidrug resistance (MDR) 4,5. Presently, identifying new antibiotics is no longer a priority for the
pharmaceutical industry and this lack of new active drugs against highly or multi-resistant microorganisms means that most infectious diseases could become untreatable, with lethal consequences in a near future 6.
During the last decades, the efforts on combating (multi)drug-resistant microorganisms were mainly focused on Gram-positive bacteria. This provided several potential novel antibiotics that, at least combined and prolonged in time, can still ensure efficacy of therapies. However, infections caused by Gram-negative bacteria are now becoming a more threatening problem mostly due to the lack of alternative treatments. The group of MDR nonfermeters causes particular concern within Gram-negative bacteria, since it represents the multi-drug resistance to the extreme 7. For the most part, Gram-negative nonfermenters
are niche pathogens able to cause opportunistic infections, for which there are few therapeutic options, due to the multidrug resistance mechanisms recurrently detected in these bacteria. Members of this group, like Pseudomonas spp. and Acinetobacter spp. are ubiquitous in nature and easily adaptable to most environmental challenges 7,8.
Mechanisms of antibiotic resistance
Bacterial antibiotic resistance can essentially be distinguished as intrinsic or acquired. Intrinsic mechanisms are those specified by naturally occurring genes, generally found on the chromosome, which allow all members of a species to resist a particular drug. On the other hand, acquired mechanisms rely on the transfer of resistance determinants born on mobile genetic elements between bacteria, even from different species. These resistance determinants are generally gene(s) that encode proteins able to protect antimicrobial targets or enzymes involved in antibiotic’s degradation, resulting in antimicrobial resistance 5,9.
In contrast to intrinsic resistance, which developed in natural populations before the widespread use of antibiotics, acquired resistance is a recent event in the evolution of human pathogens in which the main selective force has been the human use of antibiotics. Antibiotic resistance acquired by horizontal gene transfer (HGT) enables the rapid spread of resistance mechanisms from environmental bacteria to human pathogens 10.
Three main strategies of antibiotic resistance against the most use classes of antibiotics can be defined: prevention of drug access to the targets, antibiotic enzymatic inactivation and modification of the antibiotic target 4. Preventing drug access to the targets can be achieved
through, modification of permeability barriers and/or active efflux at the level of cell surface. Antibiotics must reach their specific bacterial targets and accumulate at certain quantities to be effective. When antibiotics are prevented from reaching intracellular lumen or pumped out, by drug efflux pumps, intracellular concentrations are kept low and ineffectual 4. Active
efflux pumps exist in the cytoplasmic membrane of both Gram-positive and Gram-negative bacteria. An outer membrane additionally limits Gram-negative bacteria, contributing to increase their control over diffusion of antimicrobial drugs. Exchanges across this barrier are achieved through porin channels, whose expression can be influence by mutations leading to more resistant bacteria 11.
Some microorganisms produce enzymes capable of inactivating or modifying antibiotics. Inactivation can be achieved through antibiotic hydrolysis, like β-lactamases that hydrolytically cleave the β-lactamic ring. It can also be achieved by chemical modification of the drug (phosphorylation, acetylation or adenylation) that diminishes or abolishes its activity. These enzymes are present not only in the genome of antibiotic-producing bacteria, but also on chromosomes or plasmids in bacteria from a variety of environments 12.
Modification of the antibiotic target can occur by mutations in the genetic sequence encoding that protein, hence originating an altered structure. Resistance can also be attained by changes in target concentration or by the production of proteins that protect the target form the antibiotic’s action 9,13.
Evaluation of microorganism’s susceptibility to antibiotics
The determination of minimum inhibitory concentration (MIC) is considered the ‘gold standard’ method to assess the susceptibility of microorganisms to antibiotics 14. The MIC
value corresponds to the lowest concentration of antibiotic (in mg/L) that prevents the growth of bacteria, within a defined period of time and conditions 2. Clinical breakpoints (used to
guide therapy) are based on MIC assessment and discriminate bacteria in three categories of resistance phenotype: susceptible, intermediate or resistant. Isolates are considered susceptible if their growth is inhibited by the achievable concentrations of antimicrobial agent when the recommended dosage is used for the site of infection. The intermediate category includes isolates with measured MIC’s that approach attainable blood and tissue antibiotic concentrations and for which response rates may be longer than for susceptible isolates. Resistant isolates are the ones not inhibited even by the permitted maximum dose of a given antibiotic 2.
Outside clinical settings, when the term susceptible is used in the microbiological sense, it applies to bacteria belonging to the most susceptible subpopulation that lacks resistance mechanisms. Microbiological resistant microorganisms are those possessing some resistance mechanisms demonstrated either phenotypically or genotypically 2.
Antibiotic resistance in the environment
Antibiotic resistance research has mainly been focused in clinical and veterinary settings, in contrast, the function of antibiotics in nonclinical environments has received little attention 15.
The common perception is that antibiotic resistance is manly associated with the use and overuse/misuse of antibiotics in humans and animals therapy (or prophylaxis). This is true when considering the clonal dissemination of pathogenic bacteria carrying resistance mechanisms, due to mutational events and strong selective pressure. However, the majority of antibiotic resistance mechanisms are probably transmitted through horizontal gene transfer between ecologically and taxonomically different bacteria 16.
From the microbial point of view, all sources of selective pressure contribute to resistance development so its emergence may thus come from various origins. There is an increased recognition of the importance of resistance reservoirs, which may reside in both pathogenic and non pathogenic bacteria in different ecosystems 17. According to Baquero et al. genetic
reactors are environments in which, due to the presence of specific selection, high biological connectivity and generation variation, genetic evolution of antibiotic resistance takes place. One of these defined genetic reactors is the soil and surface and ground water, where human and animal microbiota and bacteria from clinical setting, mix and counteract with environmental organisms 18.
The soil is a very rich and complex environment. Rough calculations estimate that one gram of soil holds 107 to 109 bacteria, comprising 4000 to 10000 distinct species 19. Environmental
bacteria are not globally under the strong selective pressure suffered by human and animal pathogens during antibiotic therapy. So, to properly analyse antibiotic resistance in natural ecosystems, we need to understand the function of antibiotics and antibiotic resistance genes in natural habitats. Soil bacteria and fungi naturally produce antibiotics to ensure survival in the competitive struggle for nutrients 16. Therefore, the common presence of resistance genes in antibiotic producing bacteria, or in bacteria co-sharing similar niches with the last ones, is an understandable mechanism of self-protection. In environments with little or no exposure to antibiotics, resistance determinants might nevertheless still be selected in the frame of other cellular purposes (like metabolism or signalling) 20.
In recent studies done on spore-forming bacteria from soil samples, scientists found that all strains analysed were multi-resistant, being resistance even to synthetic antibiotics, like fluoroquinolones. Despite the lack of detectable previous exposure to fluoroquinolones, 11% of the strains proved to be resistant to ciprofloxacin. They showed an high incidence level of mutations in the QRDR region, in agreement with previous studies describing natural sequence variations within this domain in soil bacteria 21,22. The soil could thus serve as an
under recognized reservoir for resistance, that has already emerged or contains enough genetic potential to emerge, with impact in clinically important bacteria. Consequently, understanding the resistance determinants present in the soil – the soil resistome – will provide information about antibiotic resistance frequencies and also about new mechanisms that may emerge in the future causing clinical problems 21.
Presence of antibiotics in environmental settings
Antibiotic concentrations in natural ecosystems increased in consequence of anthropogenic activities; the soil near animal production farms, treated with manure and contaminated water can contain antibiotics at much higher concentrations than usually found in natural environment. This might influence the selection of antibiotic resistance in microorganisms, but also alter the structure of the natural microbial populations. So these areas, contaminated with antibiotics and human associated microorganisms, are potential problematic areas that need to be studied when addressing this problem 23.
Antibiotics released in the environment can be considered pollutants. Many antibiotics are natural biodegradable compounds, but synthetic ones, like quinolones, are more resistant to biodegradation in the environment. This leads to sustained effects in time on the bacterial communities and a strong impact on resistance behaviours. Even when antibiotic contamination is cleared, resistance determinants can still be maintained in natural population since they are only lost when conferring a fitness cost to the recipient bacteria 23.
Agricultural and animal breeding practices have a major impact on the distribution of resistance elements in the environment. Antibiotics are used in agriculture as biocides and in animal breeding for veterinary medicine or as growth-promoters in animal feeds. Of more than 1 million tons of antibiotics released in the biosphere during the last 50 years, 50% are estimated to proceed from agricultural and veterinary applications. This soil pollution was shown to contribute to antibiotic resistance development and dissemination by numerous pathways 24.
The evidence is now clear that the environment should be a vast reservoir of resistant microorganisms and their associated genes. The resistome consists of all antibiotic resistance genes, including those circulating in pathogenic bacteria, antibiotic producers and benign non-pathogenic organisms, found either free living in the environment or as commensals of other organisms. This resistome is part of the incubator of the global microbial population and precedes the human use of antibiotics. The question then arises, does the environment resistome interacts with the resistome of pathogenic bacteria or are they distinct? Most resistance genes found in pathogenic bacteria are acquired thought horizontal gene transfer. This is the case of the recently discovered qnrA gene, originated from an environmental reservoir and now globally distributed in both pathogenic and non-pathogenic bacteria. The knowledge of antibiotic resistance mechanisms present in environmental bacteria will be essential in predicting the emergence of resistance and its spread providing clinicians, regulators and drug discoverers with essential information for the management of current and future antibiotics use 19.
Quinolones / Fluoroquinolones
Quinolones comprise a relatively large and expanding group of synthetic compounds. The first quinolone used, nalidixic acid, was discovered in 1962. Since then, many new compound have been developed based on a 4-oxo-1,8-naphthyridin-3-carboxylic acid; these compounds are generically known as 4-quinolones. The addition of a fluoro-group to the ring structure created a new group of quinolones, the fluoroquinolones; this is the only group of synthetic agents able to rival β-lactams impact in clinical practice. One of the most successful and widely used compounds of this class is ciprofloxacin, it entered the market in 1986 and ever since the value of fluoroquinolones for the treatment of a wide range of infections has been recognized 25-27.
Quinolones bactericidal effect results from the stable interaction complex they form with type II topoisomerases and cleaved DNA. This interaction inhibits replication and transcription of the bacterial DNA eventually leading to cell death 28. DNA gyrase and
topoisomerase IV are two essential forms of type II topoisomerases. DNA gyrase is encoded by gyrA and gyrB genes and the genes coding for topoisomerase IV are parC and parE 29.
There seems to be a pattern in which DNA gyrase is the primary quinolone target in Gram-negative bacteria and topoisomerase IV is the primary drug target in Gram-positive bacteria. The more sensitive of the two enzymes determines the drug major target 30.
As a normal part of their reaction, these topoisomerases introduce a pair of staggered single strand breaks (“nicks”) into DNA and become covalently bound to the 5’ ends of the cleaved DNA. Quinolones bind rapidly to the enzyme-DNA complex, probably before DNA cleavage occurs, trapping the bacterial II topoisomerases on DNA. The resulting structure has been termed cleavable, cleavage or cleaved complex 28,31. In consequence of cleaved
complex formation the DNA replicon machinery becomes arrested at blocked replication forks leading to inhibition of DNA synthesis. This inhibition correlates with bacteriostatic concentrations of quinolones and is thought to be reversible; DNA breaks induce the SOS regulon, which in E. coli leads to cell division inhibition and cell filamentation 28,31.
The inhibition of DNA synthesis causes slow bacterial death but evidences indicate that other hypotheses are needed to explain the rapid cellular death caused by quinolones. Studies showed that preventing the induction of the SOS system enhances bacterial death by quinolones. Other studies showed that the protein synthesis inhibitor chloramphenicol inhibits quinolone’s ability to kill bacteria. These results indicate that there is relationship between the primary effects of cleaved complex formation and the response of bacteria (through the stress-induced expression of proteins) to these effects in the bactericidal activity of quinolone antibiotics 3. According to one hypothesis quinolone’s lethality can be described
as a two-step process in which the first step is the reversible formation of cleaved complexes. This step blocks bacteria DNA replication, induces SOS response and leads to cell filamentation. In the second lethal step, requiring high ciprofloxacin concentration, DNA brakes are released from containment leading to chromosome fragmentation. This release occurs by at lest two processes, one requires protein synthesis and the other does not 28.
Quinolone resistance mechanisms
Resistance to quinolones has been described as mainly mediated by chromosomal mutations that either alters the targets or activates expression of multidrug efflux pumps for which quinolones are substrates. However, novel plasmid encoded mechanisms have been discovered recently and are now recognized as an important contributor to quinolone resistance 32.
Modification of the target enzymes arises from spontaneous mutations in the respective
encoding genes. In resistant bacteria, amino acid alteration in the GyrA and ParC subunits are usually confined to a region at the enzyme N-terminus. Mutations are less common in the gyrB and parE genes 33. Alterations described in the gyrA of E. coli are predominantly located in the so called ‘Quinolone Resistance Determinant Region’ (QRDR), between position 67 and 106 bp. A single mutation in the QRDR usually results in high level resistance to nalidixic acid. But to obtain high levels of resistance to fluoroquinolones other mutations need to induce amino acid changes in the secondary target enzyme [32, 33]. A genetic diversity study, done in environmental bacteria, showed that their QRDR is highly variable. This might, in part, explain the observed reduced susceptibility to quinolones of soil bacteria, even without apparent positive selection 22.
To reach their targets in Gram-positive bacteria, quinolones must cross the cell wall and the cytoplasmatic membrane, in Gram-negative bacteria they also need to cross the outer membrane. The cell wall doesn’t represent a major barrier to passive diffusion since quinolones are small molecules. Quinolones cross the outer membrane through specific porins or by diffusion through the phospholipid bilayer. Thus Gram-negative bacteria can regulate their outer membrane permeability by reducing the expression of porins making them more resistant to some quinolones 30,32,34.
Multiple mutations are generally required in order to obtain a clinically significant quinolone resistance level. Since spontaneous mutations are rare events, the traditional understanding of quinolone resistance as a mutational phenomenon has not been proven a fully satisfying explanation for the frequency with which this resistance has arisen. Before the discovery of quinolone resistance elements in plasmids, their existence was already suspected since horizontal transferable elements could explain how organisms survive in the presence of quinolones, while mutation occurred sequentially rather than simultaneously. Transferable elements would also explain the recurrently observed association between quinolone resistance and resistance to other antibiotics 34.
In 1998, the first plasmid mediated quinolone resistance mechanism (PMQR) was discovered when Martínez-Martínez and colleagues where studying pMG252, a plasmid from a multi-resistant strain of K. pneumoniae 35. They found a gene conferring quinolone
that Qnr acts by protecting DNA gyrase from quinolone’s interaction. Qnr belongs to the pentapeptide repeat protein family, defined by a series of tandem five aminoacids tandem repeats. After this discovery, similar proteins have been identified, and the first renamed QnrA, and the following, until now, QnrB, QnrC, QnrD and QnrS. For consensus in nomenclature qnr was defined as a naturally occurring allele encoding a pentapetide repeat protein that confers reduced susceptibility to nalidixic acid or fluoroquinolones. qnr families (such as qnrA) are defined by a 30% or less differences in nucleotidic sequence or derived amino acids 36. Within each qnr family, alleles differ in one or more amino acid. qnr genes
have been found on plasmids of different sizes and incompatibility groups, indicating that the spread of multiple plasmids has been responsible for the dissemination of this resistance around the world. Yet, they are similar enough to suggest a limited number of acquisitions followed by transposition, recombination, replication, fusion, resolution, deletion and/or insertion of DNA, to generate the diversity of plasmidic structures seen today 34. Qnr proteins
have also been found on bacterial chromosomes. In a study, where different Gram-negative bacteria were screened for qnr A-like genes, environmental Shewanella algae, was identified as the a qnrA reservoir 37.
The mechanisms through which Qnr proteins exert a protective effect against quinolone’s action are still not completely understood. It has been show that QnrA can bind specifically to the gyrase or topoisomerase IV holoenzyme, as well as to its respective subunits 38,39. This
binding occurs in the absence of relaxed DNA, ciprofloxacin or ATP, indicating that QnrA does not require the presence of the ternary complex DNA-ciprofloxacin when binding DNA topoisomerases. Whether QnrA binding blocks the access of quinolone to the enzyme, reduces the number of targets for quinolone inhibition, or as some effect on enzyme function is currently under study 38,39.
Additional PMQR mechanisms were recently found. Aac(6’)-Ib-cr is a variant of an aminoglycoside acetyltransferase which inactivates antibiotics by acetylation. Ciprofloxacin and norfloxacin are the only fluoroquinolones susceptible to such alteration so the presence of this enzyme confers selective resistance. Two different plasmid mediated quinolone transporters have also been found, OqxAB and QepA. OqxAB is a multidrug efflux pump belonging to the resistance/modulation/cell division family (RND). The qepA gene encodes a protein putatively belonging to the 14-transmembranar-segmented major facilitator superfamily of transporters 34.
Resistance mediated by Gene location / source Mechanism(s) Resistance phenotype GyrA/GyrB
ParC/ParE Chromosome Target alteration Quinolones
Efflux pumps Chromosome
/plasmids Active efflux Multiple antibiotics Porins Chromosome Decreased uptake Multiple antibiotics Qnr determinants Plasmid Target protection Quinolones
AAC(6’) – Ib - cr Plasmid Antibiotic
modification
Ciprofloxacin and norfloxacin
QepA and OqxAB Plasmid Active efflux Multiple antibiotics
Table 1. Quinolone resistance mechanisms
β-Lactams resistance
As refereed above, an association can be established between quinolone resistance and resistance to other antibiotic classes that was not explainable before the PMQR discovery. Now, different studies revealed that this association is explained by the presence of resistant determinants to other antibiotics, especially β-lactams, in plasmid carrying qnr genes. Epidemiological surveys indicate an high association rate between Qnr-positive and ESBL-positive isolates, particularly in Enterobacteriaceae 34,40.
β-lactams induce bacterial cell death by targeting cell wall metabolism. This class of antibiotics gathers the most widely used antibiotics because of their effectiveness, low cost, easy delivery and minimal side effects for humans and animals. It includes penicillin’s and derived substances, first to fifth-generation cephalosporins, monobactams and carbapenems; their distinctive structural feature is the highly reactive beta-lactam ring 41.
Bacterial cell wall is composed of a cross-linked peptidoglycan net that confers cell shape and resistance; peptidoglycan basal units are acetylmuramic acid (NAM) and N-acetylglucosamine (NAG). The reaction that links peptidoglycan units occurs outside the cytoplasmic membrane and is performed by transpeptidases (also called PBPs for Penicillin Binding Proteins). β-lactams are stereochemically similar to the penultimate D-Ala-D-Ala branch of the pentapeptide attached to NAM; hence transpeptidases mistakenly use the antibiotic as a substrate for cell wall synthesis causing the enzyme to become acylated, thus unable to hydrolyze the β-lactam. Subsequent steps in cell wall synthesis are hindered while autolysis by cell wall degrading enzymes continues. The result is a weakly linked peptidoglycan layer which makes growing bacteria susceptible to cell lyses and death 41,42.
Resistance to β-lactams can occur by three major processes: production of β-lactamases that degrade or modify β-lactams, preventing drug access to the internal target and/or target
modification (PBPs alteration). In Gram-negative bacteria, resistance is mostly due to a combination of endogenous acquired β-lactamases, and naturally up-regulated impermeability and efflux 42,43.
β-lactamases can be classified based on the enzyme functional characteristics/primary structure (Bush-Jacoby- Medeiros Groups 1 to 4) or by similarity in protein sequence (Ambler classes A to D) 44. Third generation cephalosporins were considered, when introduced in
clinical practice, as the solution for infections caused by β-lactamase producing microorganisms. But, a few years after their introduction, enzymes capable of hydrolyzing cephalosporins were reported. This new class of β-lactamases was named Extended-Spectrum β-Lactamases (ESBL). There is not a global consensus, but this class can be defined as a group of β-lactamases capable of hydrolysing penicillin, first -, second-, and third-generation cephalosporins, and aztreonam (but not cephamycins or carbapenems). ESBLs are a major therapeutic challenge because not only they are capable of degrading a wide range of β-lactams, but, in addition many ESBL producing organisms exhibit co-resistance to other antibiotic classes 43,45. Carbapenems are the treatment of choice, and the
last resort, for serious infections caused by ESBL producing bacteria. However, enzymes capable of hydrolysing carbapenems named carbapenemases have increasingly been detected and are considered an emerging treat. Carbapenemases are the most versatile lactamases, they are able to hydrolyse carbapenems but also recognize all hydrolysable β-lactams. Carbapenemases belong to two distinct molecular families, the first characterized by the presence of a serine in the active site and inhibited by clavulanic acid and tazobactam; while the second group has a zinc atom in the active site and are inhibited by EDTA 46.
Proteomics and antibiotic resistance research
Proteomics can be defined as the study of the proteome of cells, tissues or organisms and also their interaction with the environment. The proteome of an organism is the complete set of proteins it produces 47. Proteomic research is directed towards understanding protein
expression and function under specific physiologic conditions, this research can be divided into two main strategies. The first one is ‘expression’ or ‘quantitative regulation’ which consists in monitoring the expression of large number of proteins within a cell and examining how the protein expression pattern changes under different conditions. The other strategy is the ‘cell map’ or ‘structural proteomics’, here the goal is to identify the structure of proteins and how different proteins interact with each other 48.
The study of complex mixtures of proteins is mostly done by two dimensional gel - based electrophoresis (2D gels) and mass spectrometry. In 2D gels proteins are separated
according to their isoelectric points (pI) and molecular weight (mw). Then proteins can be excised form these gels and identified by mass spectrometry 47. These two classical
methods, in the field of proteomics, can be use for protein expression profiling which are quantitative catalogues of proteins produced by cells under certain conditions. Proteins from organism exposed to different stimuli are resolved on 2D gels, these gels are subsequently analysed in order to indentify the changes in protein expression in different environmental situations. So protein expression profiles can be used to compare differences in proteomes of different organisms or differences in protein expression of a single organism under different stimuli 47,49.
Genome sequencing generates genetic ‘maps’ of potential protein products and predicts functional pathways. Despite the importance of this information, the genome is ‘static’, and it does not present changes when environmental stimuli vary. On the other hand the proteome of a cell is dynamic and rapidly responding to changes in environmental conditions; it reflects the immediate surroundings in which the cell is being studied. Because changes in the proteome can reflect changes in growth conditions, exposure to specific agents or the present of mutations, protein profiles can be useful when comparing how microorganisms adapt to outside stimuli. So expression profiles techniques are being used more often to study how bacteria respond to antibiotic stress, and the mechanisms of defence involved in that process 50-52. A recent study use proteomics technology to elucidate the complex cellular
response of B. subtilis to various antimicrobial compounds, results suggest that protein expression profiles gives new insights into the bacteria response to antibiotic and could also be very useful in the process of antibiotic drug discovery 53.
AIMS
The general goal of this work was to isolate and characterize bacterial strains with significant resistance levels to quinolones, through the study of an environmental collection of bacteria sampled from soil and screened for tolerance to ciprofloxacin and nalidixic acid, from areas differently impacted by anthropogenic activities such as farms and river shores. First, isolates were characterized in terms of their resistance level to ciprofloxacin, by determination of MIC, and screened for the presence of qnr genes by mutiplex PCR.
Secondly, a selected group of isolates based on their identification and multi-resistance to ciprofloxacin and β-lactams, was used to assess global cellular response to stress induced by ciprofloxacin. This study was done by determining the impact of this antibiotic on bacterial growth curves and cellular viabilities. In addition, whole cell protein profiles were performed to compare the cells response to ciprofloxacin and as a starting point for proteomics approach on how ciprofloxacin affects cells and the intracellular pathways it triggers.
MATERIALS AND METHODS
Bacterial strainsThe bacterial strains used in this work were isolated previously from soil samples collected in a range of sampling spots in two types of locations, fluvial shores in Lisbon area and in traditional animal production farms in Northeast Portugal. In order to screen for bacterial strains potentially resistant to (fluoro)quinolones, samples were resuspended in sterile water (10 g of samples in 100 mL of H2O) and the liquid phase recovered after incubation with
agitation. The supernatant containing the extracted microbial community was then serially diluted and different concentrations plated on rich media, Luria-Bertani (LB) agar, supplemented either with ciprofloxacin or nalidixic acid, for screening isolates resistant to these antibiotics. Antibiotic where at a concentration estimated as a average breakpoint, by default, for most Gram-negative bacteria, according to The European Committee on Antimicrobial Susceptibility Testing1*.
Distinct individual colonies were collected, to enforce non-clonality of final strains within the same sampling spot, and isolated in three successive rounds of plating replications, from each sampling location. From that first screening process, 122 isolates resistant to (fluoro)quinolones were recovered. Isolates 1 to 39 grew in the presence of 1 μg/ml of ciprofloxacin, while isolates 40 to 122 were able to grow with 16 μg/ml of nalidixic acid. Strains were conserved in glycerol stocks at -80ºC. Before use in this work, bacteria were plated in LB agar and re-isolated from one single colony to ensure working with a pure culture. Then they were preserved using a commercial bead conservation system (Cryobank) and stored at -80ºC as a second stock.
The selection of Gram-negative bacteria was done by testing isolates growth on OXOID MacCONKEY AGAR No. 3 medium, overnight incubation at 37ºC.
Antibiotic susceptibility studies
Antibiotic susceptibility was determined by two techniques, broth microdilution and agar diffusion, according to the methodologies established by the Clinical and Laboratory Standard Institute 2*. The first technique enables the determination of the Minimum Inhibitory
Concentration (MIC) of the antibiotic for the tested isolate. The second one is based on antibiotic diffusion from a disc on solid inoculated growth media. The inhibition zone diameter is the area free of growth in a bacterial lawn which results from the inhibitory effect of the diffused antibiotic into the medium. In this study, susceptibilibilities to ciprofloxacin and ceftazidime were determined by broth microdilution. Susceptibility to other antibiotics,
1* http://www.eucast.org 2* http://www.clsi.org
imipenem (10 μg), meropenem (10 μg), cefpodoxime (10 μg), cefpodoxime + clavulanic acid (10 μg/1 μg) was determined by disc diffusion.
The standard inoculum for both methods was obtained by inoculating the isolates (from an overnight growth on LB agar) in 5 mL of Mueller-Hinton broth. Following 6 h of incubation at 37ºC, OD600nm was measured and dilutions were made in order to obtain a standard inoculum
corresponding to OD600nm= 0.1 or 0.5 McFarland (2x 108 cfu/ml).
To determine the MIC, serial 1:2 dilutions were performed in a 96 well microplate, with a range of eight dilutions for each antibiotic, wells were inoculated with 10 μl of standard inoculum solution. In the disc diffusion protocol, plates with 20 mL of MH agar were plated with 50 μl of standard inoculum solution and the antibiotic discs were placed on the agar surface. Inhibition zone diameters were measured after an overnight incubation at 37ºC. For both methods P. aeruginosa ATCC 27853 and E. coli ATCC 25922, controls strains for antibiotic susceptibility testing, were used according to EUCAST recommendations.
Identification of isolated strains
A preliminary triage was performed integrating data from TSI (Triple Sugar Iron medium), oxidase and catalase tests. Final identifications were obtained using API® 32GN test strips coupled with an automated system and software (BioMérieux).
The identification of the strains used on the final studies was confirmed by 16S rRNA gene amplification and sequencing. The primers used were the following: 27F: 5’AGAGTTTGATCMTGGCTAG3’ and 1492R: 5’GYTACCTTGTTACGACTT3’. Amplification was carried out with the following thermal cycling profile: 5 min at 95ºC and 35 cycles of amplification consisting of 1 min at 95ºC, 1min at 50ºC and 3 min at 72ºC and 10 min at 72ºC for the final extension. Amplification was performed using a proof reading enzyme PFU DNA polymerase (Fermentas). PCR products were verified in a 2% agarose gel and then concentrated and precipitated before being sent to sequentiation by STABVida 54.
S. maltophilia and ciprofloxacin
Growth curves and cellular viability
S. maltophilia growth curves were performed by measuring the culture optical density (OD600nm) over time (Evolution 300 UV-Vis, Thermo Scientific). All growth curves were
initiated by diluting a pre-inoculum (overnight growth in LB broth) in order to generate a starter culture in 50 mL (LB broth) at OD600nm ≈ 0.1. To test how ciprofloxacin affects S.
maltophilia growth, isolates growth curves were performed in the seven following conditions: Control (without antibiotic), upon addition of ciprofloxacin as half its MIC (½ MIC exp, ½ MIC stat), as its MIC (MIC exp, MIC stat) and at twice its MIC (2xMIC exp, 2xMIC stat). All
cultures were incubated in an orbital incubator at 37ºC and 120 rpm of agitation (Innova 40 New Bronswick Scientific).
Simultaneously, analysis of cellular viability was performed. After addition of ciprofloxacin (60min) a fraction of 100μl of the culture was collected. In a microplate, this culture was diluted in series of 10-2 to 10-10, and then 5 μl of each dilution inoculated on LB agar and
incubated overnight at 37ºC.
Protein profiles
Bacterial cells were collected for protein extraction 30 and 60 min after ciprofloxacin was added to the growing culture in order to analyze cell total protein profiles variations. A total of 3 mL of culture was collected, after centrifugation (5000 g for 2 min) the supernatant was discarded and the pellets store at -20ºC. Protein extraction was performed by ressuspending cells in 500 μl of lysis buffer (SDS +Tris) and bacteria were disrupted by ultra-sonication (5x5s, 20 kHz). The resulting solution was centrifuged to remove cell debris and the supernatant containing the total protein extract stored at -20ºC. Protein concentration was estimated by OD280nm measurements using a Nanodrop spectrophotometer.
Whole cell protein extracts were analysed by ‘Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis’ (SDS-PAGE). Dilutions were made from protein solutions to obtain 10μl samples containing 40 μg of total proteins to which were added 10 μl of a 10X migration dye and β-mercaptoethanol solution. Before application on the gel samples were boiled for 3 min to ensure full denaturation of the proteins. We used discontinuous polyacrylamide formed by a 5% stacking gel and a 15% running gel. Electrophoresis was performed in denaturating conditions at constant amperage (20 mA) in a Mini PROTEAN Tetra System (Bio-Rad). Gels were stained with a Coomassie blue solution (0.1% Coomassie Brilliant Blue, 50% methanol, 10% acetic acid) for at least 1 h or overnight. Gels were incubated in a distaining solution (30% ethanol, 10% acetic acid) for 1 to 2 h. Electrophoresis in larger polyacrilamid gels were performed at 4ºC and the total protein concentration loaded per lane was 150 μg all the other conditions were as used in the mini gels, adapting volumes of polyacrilamide and buffers.
Total DNA extraction
Cellular pellets collected in exponential bacterial growth were resuspended in 525 μl of milliQ water and mixed with 15 μl of 20% SDS and 60 μl of STEP solution (0.5% SDS 50 mM, Tris-HCl pH 8, 0.4 M EDTA, 1 mg/ml proteinase K) to lyse cells. After 1 h of incubation at 37ºC plus 30 min at -80ºC an equal volume of phenol/chloroform pH 8 was added and then mixture homogenized. Once centrifuged the supernatant was recovered and precipitate with 1/10 volume of 3 M sodium acetate and 2.5 vol. of ethanol 100% and stored for 30 min at -80ºC. The precipitate was centrifuged for 20 min at 4ºC and 5000 g and the supernatant
was discarded. The pellet was washed twice with 70% ethanol. After drying, the pellet was ressuspended in 1 mL of milliQ water. DNA quality was assessed by 2% agarose/TBE 1X gels and, when needed, DNA was treated with RNase A to remove RNA contaminants. To 50 μl of DNA solution 2,5 μl of 1:20 solution of RNase A was added, after 30 min of incubation at 37ºC 100 μl of phenol: chloroform solution was added. Following centrifugation, the supernatant was transferred and DNA precipitated as referred before. DNA concentration was measured at OD260nm using a Nanodrop spectrophotometer.
Screening for Qnr-positive isolates
The detection of qnr genes (qnrA, qnrB and qnrS) was performed by multiplex PCR using the primers described in Cattoir et al. 55. PCR reactions were performed using a commercial
reaction mix (BioMix from Bioline), three specific sets of primers and total DNA. Amplification was carried out with the following thermal cycling profile: 10 min at 95ºC and 35 cycles of amplification consisting of 1 min at 95º C, 1 min at 54ºC and 1 min at 72ºC and 10 min at 72ºC for the final extension. PCR products were analyzed in 1.5% agarose/ TBE 1x gels.
Primers Sequence (5' - 3') Gene Size of the amplicon
A1(F) A2(R) AGAGGATTTCTCACGCCAGG TGCCAGGCACAGATCTTGAC qnrA1 to qnrA6 580 bp B1(F) B1(R) GGMATHGAAATTCGCCACTG TTTGCYGYYCGCCAGTCGAA qnrB1 to qnrB6 264 bp S1(F) S2(R) GCAAGTTCATTGAACAGGGT TCTAAACCGTCGAGTTGCGGC qnrS1 to qnrS2 428 bp
Table 2. Sequence and amplification product of the primers used in the qnr multiplex
PCR. M=A or C; H= A or C or T; Y= C or T 55
RESULTS AND DISCUSSION
Environmental isolates resistant to (fluro)quinolones
In this work, a collection of 122 environmental isolates recovered from agar plates supplemented with ciprofloxacin or nalidixic acid was studied. This collection was composed of 39 isolates obtained by selection with ciprofloxacin and 83 obtained by selection with nalidixic acid. Analyses of the QRDR of gyrA gene revealed that this sequence is highly variable in natural microbial population 22. Single mutations in the QRDR region usually
results in high resistance levels to primordial quinolones (as nalidixic acid), but for fluoroquinolones (ciprofloxacin) resistance additional mutations or mechanisms are usually required. Thus it is simpler to acquired resistance to nalidixic acid (one step) than to fluoroquinolones (two or more steps), which probably explains the prevalence of bacteria isolated through the isolation screening with acid nalidixic in this collection.
Selection of Gram-negative bacteria
Our focus was on the analysis of Gram-negative bacteria and OXOID McCONKEY AGAR No. 3 medium was thus used to ensure that selection since it contains bile salts and crystal violet, growth inhibitors of Gram-positive bacteria. Amongst the 122 isolates, 10 were unable to grow on McConkey agar, isolates 15 to 17, 20 to 24, 35 and 37, therefore considered Gram-positive bacteria and removed from the bacterial collection to be studied. This collection of 112 environmental Gram-negative bacteria was subsequently used.
Determination of ciprofloxacin MIC
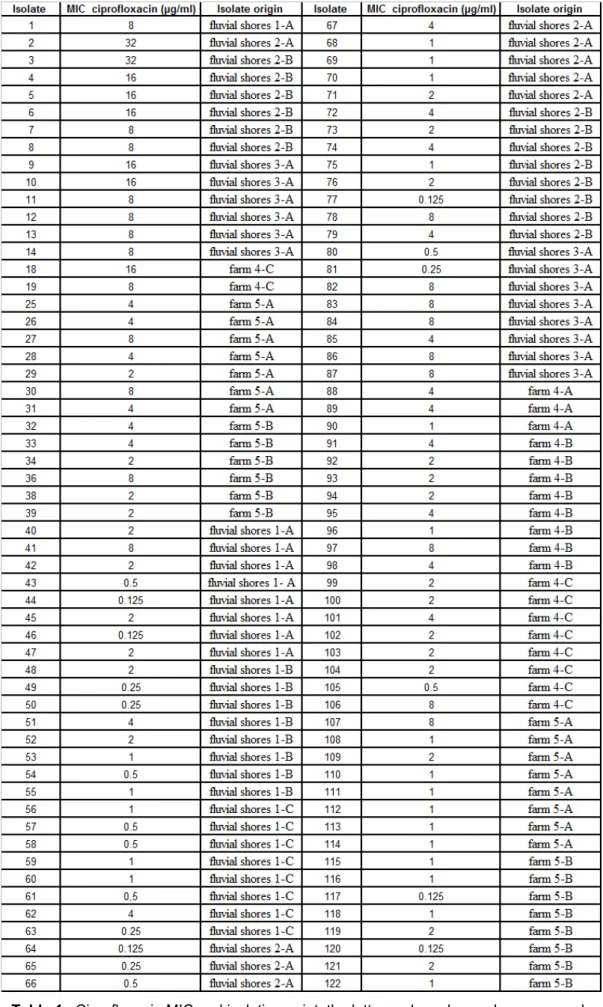
With a total of 112 Gram-negative resistant bacteria, the starting point was to determine their ciprofloxacin MIC. The chosen method was broth microdilution, which was implemented and optimized in the context of this work, and performed at least in triplicates for each isolate. Results are shown in annexed Table 1 and along with the ciprofloxacin MIC are also provided the sampling locations. These Gram-negative resistant bacteria were isolated from soil from two distinct environments, 62 were collected in river Tejo shores and 50 recovered from animal production farms.
At this stage, without a phylogenetic analysis approach, which falls out of the scope of this work, it remains possible that some isolates are clones, since the same extracted microbial community from the soil in each sampling spot was screened with nalidixic acid and ciprofloxacin. Environmental isolation of bacteria depends on a diverse mix of factors that range from the growth medium used to the characteristics of the isolates, which increases its
complexity. For instance, farm sampling locations 4A and 4B only yielded isolates through acid nalidixic screening. However, the ciprofloxacin MIC determined for these 11 isolated strains are all ≥ 1 μg/ml, the screening breakpoint used in ciprofloxacin screening, where they should have been isolated in parallel. This shows how subjective isolation procedures can be, even varying only one parameter, such as antibiotic.
By integrating the ciprofloxacin MIC values with the information on the origins of sampling (Table 1. Annex), 6 possible replicated clones can be pointed, such as isolates 1/41 or 29/109, corresponding to a possible error of 5% in analysis of this isolates collection.
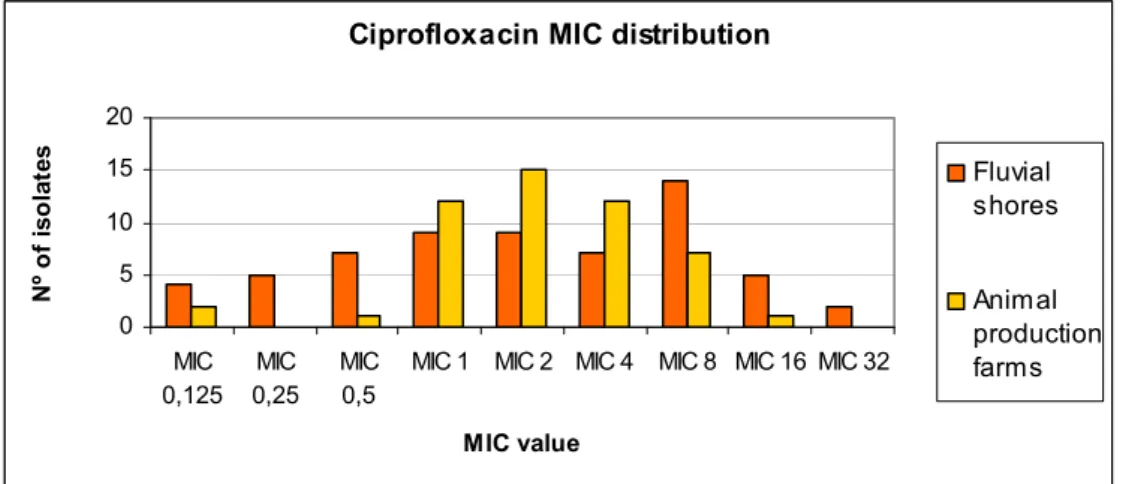
Therefore, ciprofloxacin resistance levels were compared in the in the two sampling locations (Figure 2), showing the distribution according to ciprofloxacin MIC. Some isolates with MIC values below the ciprofloxacin breakpoint used for screening (1 μg/ml) correspond to strains recovered through the screening with acid nalidixic. As mentioned above, resistance to quinolones does not imply resistance to fluoroquinolones. However, amongst the 83 isolates obtained by nalidixic acid screening, 77% are tolerant to ciprofloxacin. Only 6% of the isolates from animal production farms present ciprofloxacin MICs below 1 μg/ml, in contrast with, a more significant number (26%) of fluvial shore isolates being susceptible to ciprofloxacin. Independently of their isolation environment, the majority of the isolates (76%) obtained with this screening procedure has ciprofloxacin MICs between 1 and 8μg/ml. More resistant strains with higher MICs (≥ 8 μg/ml) are less frequent, especially in those originating from animal production farms. Most of these isolates with stronger resistance potential, around 66%, have been isolated from urban shores. Interestingly the bacteria with higher resistance level to ciprofloxacin (32 μg/ml) came from the fluvial shores.
Ciprofloxacin MIC distribution
0 5 10 15 20 MIC 0,125 MIC 0,25 MIC 0,5
MIC 1 MIC 2 MIC 4 MIC 8 MIC 16 MIC 32
MIC value N º of is ol at es Fluvial shores Animal production farms
Figure 2. Comparison between fluvial shores and animal production farms
isolates in terms of their ciprofloxacin MIC (expressed in μg/ml)
The fluvial shores represent an environment susceptible to the most diverse contaminations, including antibiotics, through urban sewage or discharges waste waters and industrial or fluvial transportation of pollution. On the other hand, soil from animal production
farms is an environment also heavily contaminated, in this case with antibiotics used in treatments or prophylaxis, biocides used for sanitary purposes and resistance microorganisms due to fecal cross-contamination from animals. Isolates with different ciprofloxacin resistance profiles were recovered from both environments, so none environments appear to favour emergence of ciprofloxacin resistance more than the other. However, it is clear that most isolates from farm origins have basal resistance levels to ciprofloxacin higher than fluvial strains, probably due to the selective process related to sustained presence of antibiotics is this locations.
Screening for Qnr-positive isolates
As referred in the introduction, the recently discovered qnr gene has a worldwide
distribution and is easily spread in environmental bacteria due to their plasmidic location. This collection of environmental isolates was screened for the presence of Qnr-positive isolates using a multiplex PCR technique, with primers designed to amplify qnrA, qnrB and qnrS. Primers designed for qnrB amplification are degenerated to overcome the variability of this group. As PCR positive controls were used three control strains, issued from the EU-CRL for antibiotic resistance (European Community, Reference laboratory) at the Danish National Food Institute, each possessing one of these qnr alleles.
Using this set of primers and the amplification conditions described in the methods for the 112 isolates, only one isolate, later on identified as Comamonas testosterone, presented positive amplification. Repeated PCR results revealed a band with 420 bp, probably corresponding to the expected size of qnrS, and an unspecific band with 320 bp; this result wasn’t conclusive to determine if this C. testosterone isolate carries a qnr gene. Additional analyses are required, namely sequencing the 420 bp amplicon and compare it with the qnr genes sequenced so far, in order to determine if this is the first qnr discovered in a C. testosterone.
Nevertheless, this screening shows a very low prevalence, or even absence, of qnr variants among the environmental isolates of this collection. Alternative mechanisms of resistance should therefore be related to the significant levels of resistance to ciprofloxacin found generally among them.
Isolates selection and identification
In the second part of this work, the main goal was to study how ciprofloxacin influences the growth and cellular viability and how this reflects on protein profiles. To perform these analyses a smaller group of isolates was selected based in a two step process: first 16 isolates presenting higher ciprofloxacin MICs were chosen for identification, secondly, these
bacteria were characterized in terms of multi-resistance behaviour through assessment of susceptibility to β-lactams.
Taking into consideration their high ciprofloxacin MIC and also the sampling spots, in order to avoid choosing clonal duplicates, the following 16 isolates were selected for identification: 1, 2, 3, 4, 5, 6, 7, 9, 11, 18, 25, 27, 29, 34, 36 and 38. First, preliminary biochemical testes were performed to establish broad strain characteristics and determine the API gallery to be used. TSI, oxidase and catalase tests inferred the ability to ferment dextrose, lactose, sacarose and sulphured compound and to produce oxidase and/or catalase. Final identification results were obtained using API ® 32 GN test strips designed for the identification of Gram-negative rods. The identification results are shown on Table 3.
Numerical identification is based on the calculation, for the observed profiles, of its relative proximity to the different taxa of the data base - identification percentage (%id), and of its proximity to the most typical profile in each of the taxa - T index (T). The quality of the identification is determined based on %id and T values. So considering these two values we can conclude that the identification of isolates 1, 3, 4, 6, 11 and 38 is an excellent identification (%id ≥ 99.9 and T≥0.75). For isolates 2, 5, 7 and 9 we have a very good identification (%id ≥ 99.0 and T≥0.5), isolates 18 and 25 have a good identification (%id ≥ 99.0 and T≥0.25); the remaining isolates had an acceptable identification (%id ≥ 80.0 and T≥0) 56.
Isolates Identification results %id / T
1 Comamonas testosteroni 99.9 / 0.75 2 Acinetobacter baumanni 97.9 / 0.63 3 Stenotrophomonas maltophilia 99.9 / 0.95 4 Stenotrophomonas maltophilia 99.9 / 0.84 5 Stenotrophomonas maltophilia 99.7 / 0.72 6 Stenotrophomonas maltophilia 99.9 / 0.95 7 Stenotrophomonas maltophilia 99.7 / 0.72 9 Stenotrophomonas maltophilia 99.7 / 0.72 11 Stenotrophomonas maltophilia 99.9 / 0.95 18 Acinetobacter baumanni 99.9 / 0.42 25 Comamonas testosteroni 97.5 / 0.44 27 Stenotrophomonas maltophilia 83.5 / 0.71 29 Comamonas testosteroni 98.5 / 0.81 34 Aeromonas sobria 83.1 / 0.42 36 Comamonas testosteroni 82.9 / 0.50 38 Acinetobacter lwoffii 99.9 / 0.84