ContentslistsavailableatScienceDirect
Systematic
and
Applied
Microbiology
j o ur na l h o me pa g e :h t t p : / / w w w . e l s e v i e r . c o m / l o c a t e / s y a p m
History
and
current
taxonomic
status
of
genus
Agrobacterium
José
David
Flores-Félix
a,
Esther
Menéndez
a,1,
Alvaro
Peix
b,c,
Paula
García-Fraile
a,
Encarna
Velázquez
a,c,∗aDepartamentodeMicrobiologíayGenéticaandInstitutoHispanolusodeInvestigacionesAgrarias(CIALE),UniversidaddeSalamanca,Salamanca,Spain bInstitutodeRecursosNaturalesyAgrobiología,IRNASA-CSIC,Salamanca,Spain
cUnidadAsociadaGrupodeInteracciónPlanta-Microorganismo(UniversidaddeSalamanca-IRNASA-CSIC),Salamanca,Spain
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received21May2019 Receivedinrevisedform 15November2019 Accepted22November2019 Keywords: Agrobacterium Taxonomy Rhizobium Allorhizobium
a
b
s
t
r
a
c
t
ThegenusAgrobacteriumwascreatedacenturyagobyConnwhoincludeditinthefamilyRhizobiaceae togetherwiththegenusRhizobium.Initially,thegenusAgrobacteriumcontainedthenon-pathogenic species Agrobacteriumradiobacter andthe plantpathogenic species Agrobacteriumtumefaciens and Agrobacteriumrhizogenes.Attheendofthepastcenturytwonewpathogenicspecies,Agrobacterium rubiandAgrobacteriumvitis,wereaddedtothegenus.Alreadyinthepresentcenturythesespeciesplus AgrobacteriumlarrymooreiwerereclassifiedintogenusRhizobium.Thisreclassificationwas controver-sialandforatimebothgenusnameswereusedwhennewspeciesweredescribed.Fewyearsago, afterataxonomicrevisionbasedongenomicdata,theoldspeciesA.rhizogeneswasmaintainedinthe genusRhizobium,theoldspeciesA.vitiswastransferredtothegenusAllorhizobiumandseveral Rhizo-biumspeciesweretransferredtothegenusAgrobacterium,whichcurrentlycontains14speciesincluding theoldspeciesA.radiobacter,A.tumefaciens,A.rubiandA.larrymoorei.Mostofthesespeciesareable toproducetumoursindifferentplants,neverthelessthegenusAgrobacteriumalsoencompasses non-pathogenicspecies,onespeciesabletonodulatelegumesandonehumanpathogenicspecies.Taking intoaccountthatthespeciesaffiliationstofiveAgrobacteriumgenomospecieshavenotbeendetermined yet,anincreaseinthenumberofspecieswithinthisgenusisexpectedinthenearfuture.
©2019PublishedbyElsevierGmbH.
TheAgrobacteriumtaxonomyinthepastcentury
The genus Agrobacterium was created by Conn [9] who includeditwithinthefamily Rhizobiaceae[8]togetherwiththe genus Rhizobium [14]. In the 5th edition of the Bergey’s Man-ualofDeterminativeBacteriology, thespecies includedintothe genus Agrobacterium were Agrobacterium radiobacter, Agrobac-teriumtumefaciensandAgrobacteriumrhizogenes.Inthe6thedition ofthisManualtheplanttumorigenicspeciesAgrobacteriumrubi [17,48]wasincluded,beingdifferentiatedfromA.tumefacienson the basis of the ability to reduce nitrate to nitrite [6]. In the 7theditionofthisManualthespeciesAgrobacteriumstellulatum, Agrobacteriumpseudotsugae andAgrobacteriumgypsophilaewere addedto thegenus[7],but this inclusionwas laterconsidered
∗ Correspondingauthorat:DepartamentodeMicrobiologíayGenética,Lab.209, EdificioDepartamentaldeBiología.CampusMigueldeUnamuno,37007Salamanca, Spain.
E-mailaddress:evp@usal.es(E.Velázquez).
1 Currentaffiliation:ICAAM–InstitutodeCiênciasAgráriaseAmbientais
Mediter-rânicas,LaboratóriodeMicrobiologiadoSolo,UniversidadedeÉvora.
unjustified becausethey probably belongto othergenera [23]. Therefore, in the subsequent 8th edition of the Bergey’s Man-ual only fourspecies were included,A. radiobacter,which was non-pathogenic,A.rhizogenes,whichinducedhairyroots,andA. tumefaciensandA.rubi,whichinducedplantgalls.The differentia-tionofthesespecieswasbasedonsomephenotypiccharacteristics togetherwiththeirabilitytoproducediversesymptomsin differ-entplants[1].Thesefourspecieswereincludedinthevalidation listsofSkermanetal.[45],whoindicatedthatA.tumefaciensisthe typespeciesofgenusAgrobacterium.
Duringthe70and80softhepastcentury,severalstudies includ-ing strainsisolatedfromdifferent sourceswere performedand theexistenceofdifferentvarieties,biovarsand/orbiotypeswas deeply revisedby Young [58].Some authorsproposedthat the speciesA.tumefaciensandA.radiobacterconstituteasinglespecies, A.radiobacter[20],laternamedA.radiobactervar.tumefaciens[21]. However,consideringthatA.tumefacienswasconsideredthetype speciesofthegenusAgrobacteriumbySkermanetal.[45],Holmes andRobertsconsideredthatthecorrectnamefortheunifiedtaxon isA.tumefaciens[18].
https://doi.org/10.1016/j.syapm.2019.126046 0723-2020/©2019PublishedbyElsevierGmbH.
Nonetheless,inthe1steditionoftheBergey’sManualof Sys-tematicBacteriologypublishsedin1984,KerstersanddeLey[22] maintainedthefourspecies,buttheyincludedthreebiovarsinthe speciesA.tumefaciensandtwobiovarsinthespeciesA.radiobacter, assigningthespeciesA.rhizogenestoanamelybiovar2and main-tainingthespeciesA.rubi.InthiseditionoftheBergey´sManual ahighernumberofphenotypiccharacteristicswererecordedfor theAgrobacteriumspecies,buttheirdifferentiationcontinuedtobe mainlybasedonphenotypicandphytopatogenicitytests[22].
Atthe beginning ofthe 90sseveral changes wereproposed withinthegenusAgrobacteriumincludingthedescriptionofnew species,neverthelessnoneofthesechangeswererecordedinthe 9thandlasteditionoftheBergey’sManualofDeterminative Bacte-riologypublishedin1994,whichincludedthesamespeciesthatthe previousversionofthisManual[19].Inyear1990thestrainsfrom thebiovar3ofA.tumefaciensproducingtumoursinVitisvinifera wereincludedintoanewspeciesofgenusAgrobacteriumnamed A.vitis[32].In thedescriptionofthisspecies,inadditiontothe classicphenotypicandphytopatogenicytests,theauthorscarried outDNA–DNAhybridizationamongthetypestrainsofthe Agrobac-teriumspeciesdescribedtodateandamongseveralstrainsofthe newproposedones.Theresultsobtainedinthisworkshowedthat thetypestrainofA.vitispresentedlessthan50%DNA–DNA related-nesswithrespecttothetypestrainsoftheremainingAgrobacterium species,whereastheanalysedstrainsofA.vitispresentedvalues rangingfrom80to92%amongthem.Surprisingly,thetypestrains A.tumefaciensNCPPB2437TandA.radiobacterATCC19358Talso
presentedDNA–DNArelatednessvalueswithinthisrange[32]. In1992,RügerandHöfle[41]includedseveralstrainsisolated frommarinesourcesinseveralspeciesofthegenusAgrobacterium recoveringthenamesA.stellulatum,Agrobacteriumferrugineumand Agrobacteriumgelatinovorum,whichwerenotincludedinthe val-idationlistsofSkermanetal.[45],andproposingthenewspecies AgrobacteriumatlanticumandAgrobacteriummeteori.Inthiswork theAgrobacteriumspeciespreviouslydescribedwerenotincluded andtheclassificationofthenewproposedoneswasbasedinthe initialassignement of A.stellulatumtothe genusAgrobacterium [47] being differentiated the newspecies onthe basis of phe-notypictraits and thelow DNA–DNArelatedness foundamong them[41].Allthesespecieswerelaterreclassifiedinothergenera. ThespeciesAgrobacteriumatlanticumand Agrobacteriummeteori werereclassifiedintoRuegeriaatlantica[51,52].Thetypestrainof Agrobacteriumferrugineumwasreclassifiedinto Pseudorhodobac-terferrugineus[50,53]andthestrainLMG128intoHoefleamarina [37].AgrobacteriumstellulatumwasreclassifiedintoStappia stellu-lata[51,52]andAgrobacteriumgelatinovorumwasreclassifiedinto Ruegeriagelatinovorans[51,52]andlaterintoThalassobius gelati-novorus[2].
Ayearafter,Sawadaetal.[43]revisedthetaxonomicstatusof Agrobacteriumspeciesdescribeduntilyear1990.Inthisworkthe 16SrRNA(rrs)genesequencesofrepresentativestrainsofthe bio-var1(A.tumefaciens-A.radiobacter),biovar2(A.rhizogenes),biovar 3(A.vitis)andA.rubiwereobtainedandcomparedwiththoseofthe severalspeciesfromdifferentalphaProteobacteriagenera, includ-ingthegenusRhizobium.Theanalysisofthisgeneshowedthatthe strainsfrombiovar1wererelatedtoA.rubi,butthosefromthe bio-vars2and3werephylogeneticallydivergentbeingthestrainsfrom thebiovar2verycloselyrelatedtothetypestrainofRhizobium trop-ici.Nevertheless,despitethespeciesA.rhizogeneswasemended,it wasmaintainedwithinthegenusAgrobacterium.Moreover,itwas reportedthatthetypestrainsofA.tumefaciensNCPPB2437TandA.
radiobacterIAM12048Thave87%DNA–DNArelatednessin
agree-mentwiththeresultsfromOphelandKerr[32]forA.tumefaciens NCPPB2437TandA.radiobacterATCC19358T.Therefore,Sawada
etal.concludedthatthesetwostrainscannotbeplacedin differ-entspeciesandtheyindicatedthataccordingtotheRule38ofthe
InternationalCodeofNomenclatureofBacteriathespecies estab-lishedearliershouldbemaintained[28].SinceA.radiobacterwas describedin1902(asBacillusradiobacter)byBeijerinckandvan Delden[4]andA.tumefacienswasdescribedin1907(asBacterium tumefaciens)bySmithandTownsend[46],Sawadaetal.[43] pro-posedtonamethestrainsofbiovar1asA.radiobacterandtoreject thespeciesnameA.tumefaciens.Thus,attheendofthepastcentury A.radiobacter,A.rhizogenes,A.rubiandA.vitiswereconsideredthe validspeciesofgenusAgrobacterium.
ThetaxonomyofgenusAgrobacteriumfrom2000onwards
Themostrelevantandconflictivechangeinthetaxonomic sta-tusofgenusAgrobacteriumtookplaceinyear2001whenYoung et al. [60] proposedthe reclassification of the complete genus AgrobacteriumintothegenusRhizobium.Thisreclassificationwas based on the resultsof Sawada et al. [43] and, therefore, the Agrobacterium species were reclassified as Rhizobium radiobac-ter,Rhizobiumrhizogenes,RhizobiumrubiandRhizobiumvitis.The reclassification,whichwasbasedontherrsgenesequences ana-lysed by Maximum-likelihood and Neighbour-joining methods, wasnotacceptedbymanyauthorswhosignedtheletterof Far-randetal.[13].Theseauthorsconsideredthatonlythebiovar2 belongstogenusRhizobiumandthatclassicalandmoleculardata supportthatAgrobacteriumandRhizobiumaredifferentgenera.This letterwaslaterrepliedbyYoung etal.[61]andthecontroversy wassettledbykeepingthereclassification.Consideringthis decis-sion,Young[57]alsoproposedthereclassificationofthespecies Agrobacteriumlarrymoorei,whichwasdescribedin2001[5],as Rhi-zobiumlarrymoorei.Nevertheless,thisspecieswasnotincludedin the2ndeditionoftheBergey’sManualwhichonlyincludedthe speciesA.tumefaciens,A.rhizogenes,A.rubiandA.radiobacter. Curi-ously,inthiseditionofBergey’sManualtheofficialreclassification ofAgrobacteriumspeciesintothegenusRhizobiumwasdisregarded, althoughtheauthorsmakeahistoricalreviewofthenomenclature problemsaffectingthegenusAgrobacteriumoverthetime[59].
Duetotheconfusiongeneratedafterthereclassificationofgenus Agrobacterium intogenusRhizobium, thenewspecies described since2001outsidetheofficialjournalfordescriptionofnewtaxa ofProkaryotes(IJSEM)werenamedAgrobacterium,asoccurredin thecaseofAgrobacteriumalbertimagniisolatedfromaquaticplants, in whose description,as occurredin the case of A. larrymoorei describedayearbefore,onlytherrsgenewasincluded[42]. How-ever,thespeciesdescribedoutside thementionedjournalwere namedRhizobiumandtheanalysisofseveralhousekeepinggenes wasincludedintheirdescriptionsin additiontothatoftherrs gene.ThesespecieswereRhizobiumpusense,isolatedfrom chick-pearhizosphere[35],Rhizobiumnepotum,isolatedfromtumoursof differentplants[38]andRhizobiumskierniewicenseisolatedfrom tumoursofchrysanthemumandcherryplum[39].
Inparallel,theAgrobacteriumstrainsfromdifferentspecieswere distributedintothegenomicgroups(genomicspecies, genomo-species,genomovars)G1toG9,G11,twounnamedgenomicgroups andG13[10,11,30].Atthetimeoftheseworks,theG4includedthe typestrainsofA.tumefaciensandA.radiobacter,theG11thatofA. rubi,theG10thatofA.rhizogenesandthetwounnamedgroups includedseveralstrainsofA.larrymooreiandA.vitis[30].Laterit wasshownthattheG8correspondstothespeciesA.fabrum[29], theG2tothespeciesR.pusense[3,35]andanewgenomovar,named G14,correspondstothespeciesR.nepotum[38,44].
Thetaxonomicstatusofthesespecieswaschangedagainwhen Mousaviet al. [31] basedon theanalysisof the rrs,recA, atpD andrpoBgenesproposedtotransferR.pusense, R.nepotumand R.skierniewicensetogenusAgrobacteriumandR.vitis(initiallyA. vitis)togenusAllorhizobium,asnewcombinations, whichwere
Table1
SpeciescurrentlyincludedinthegenusAgrobacteriumandspeciescausingtumoursorhairyrootscurrentlyincludedinothergeneraofFamilyRhizobiaceae.
Species Sourceofisolation Pathogenicitysymptoms References
GenusAgrobacterium
A.radiobactera Soilandplantrhizosphere Non-pathogenic [4,9]
A.tumefaciensa Malussp.tumours Tumours [9,46]
A.rubi Rubussp.tumours Tumours [17,48]
A.larrymoorei Ficusbenjaminatumours Tumours [5]
‘A.albertimagni’ Potamogetonpectinatus Nodata [42]
‘A.fabrum’ Prunussp.,Humuluslupulus,Euonymusalata,Rubusmacropetalustumours Tumours [29]
A.pusense Cicerarietinumrhizosphere Nodata [31,35]
A.nepotum Prunus,VitisandRubustumours Tumours [31,38]
A.skierniewicense ChrysanthemumandPrunustumours Tumours [39]
A.arsenijevicii PrunusandRubustumours Tumours [25]
‘A.deltaense’ Sesbaniacannabinanodules Nodata [55]
A.salinitolerans Sesbaniacannabinanodules Nodata [56]
‘A.bohemicum’ Papaversomniferum Non-pathogenic [62]
A.rosae Rosaxhybridatumours Tumours [26]
GenusAllorhizobium
A.vitis Vitisviniferatumours Tumours [31,32]
GenusRhizobium
R.rhizogenes Malussp. Hairyroots [9,40]
‘R.tumorigenes’ Rubussp.tumours Tumours [27]
aWehaveincludedthesetwospeciesnamestakingintoaccountthecurrentdecisionoftheJudicialCommision[49].Ganetal.[22]proposedthattheybelongtodifferent
subspeciesofthespeciesA.radiobacter.Thequotednamescorrespondtospeciesthathavenotbeenofficiallyvalidatedtodate.
later validatedin IJSEM [33]. Theold species A. rhizogeneswas maintainedasRhizobiumrhizogenesinthegenusRhizobium,which currentlyalsocontainsthenewspeciesRhizobiumtumorigenesable toinduceplanttumours[27].Mousavietal.[31]didnotconsidered thespeciesAgrobacteriumfabrumisolatedfromtumoursof vari-ousplants[29]becauseitwasnotofficiallyproposed,althoughthe completegenomeofitstypestrainC58wasthefirstobtainedfor amemberofthegenusAgrobacteriumanditwassequencedtwice independentlyduetotheimportanceofthisstrainforplantgenetic engineering[16,54].
Since2015,severalnewspeciesofgenusAgrobacteriumhave been described based on the analyses of the rrs and several housekeepinggenesand,insomecases,thecomplete genomes. Someofthesespecieswereisolatedfromplanttumours,suchas Agrobacteriumarsenijevicii[25]andAgrobacteriumrosae[26],but otherswereisolatedfromlegumenodules,suchasAgrobacterium deltaense[55]andAgrobacteriumsalinitolerans[56],orplantwastes, suchasAgrobacteriumbohemicum[62].Insomeoftheseworksa strainnamedKB-105(ATCC31113)fromAgrobacteriumviscosum wasmentioned[12],neverthelessthisspecieshasnotbeen offi-ciallyproposedandmoreoverthestrainKB-105,whichis nota type strain,iscurrently includedina patent (U.S.Patent Num-ber4,028,185).Therefore,A.viscosumcannotbeconsideredavalid speciestodateandthenithasnotbeenincludedinthepresent study.
ConcerningtotheoldspeciesofgenusAgrobacteriumthatwere nottransferredtoothergenerabyMousavietal.[31],theyreturn totheirinitialnamesA.tumefaciens,A.radiobacter,A.rubiandA. larrymoorei.Nevertheless,there isaproblemwiththenamesof thetwofirstspeciesaswaspointedoutbySawadaetal.[43]who, consideringthatthenameA.radiobacterhaspriority,proposedthe rejectionofthenameA.tumefaciens.Nevertheless, accordingto Rule56aoftheInternationalCodeofNomenclatureofBacteria,only theJudicialCommissioncanplaceanameonthelistofrejected names[28,36].Afteryearsofdiscussionaboutthisissue,thelast decisionofthisCommissiontakenin2014isthat“the combina-tionAgrobacteriumradiobacter(BeijerinckandvanDelden1902) Conn1942haspriorityoverthecombinationAgrobacterium tume-faciens(SmithandTownsend1907)Conn1942whenthetwoare treatedasmembersof thesamespeciesbasedontheprinciple ofpriorityasappliedtothecorrespondingspecificepithets.The typespeciesofthegenusisAgrobacteriumtumefaciens(Smithand Townsend1907)Conn1942,eveniftreatedasalaterheterotypic
synonymofAgrobacteriumradiobacter(BeijerinckandvanDelden 1902)Conn1942.Agrobacteriumtumefaciens(SmithandTownsend 1907)Conn1942istypifiedbythestraindefinedontheApproved ListsofBacterialNamesandbystrainsknowntobederivedfrom thenomenclaturaltype”[49].
In 2014,thesame yearthedecision oftheJudicial Commis-sionwaspublished, Zhangetal. [63]sequencedthegenomeof thetypestrainofA.radiobacterheldinDSMZculturecollection (DSM30147T)andproposedtheemendationofthisspeciesinorder
toincludetheresultsofassimilationofdifferentcarbonsources, theproductionofseveralenzymesandthefattyacidprofile.These authorsindicatedthatthegenomeofthestrainDSM30147Twas
unusuallylarge,sinceithasmorethan7Mbwhereastheremaining analysedgenomeshavesizesrangingfrom4.8to6.8Mb. Neverthe-less,inthepresentyear2019,Ganetal.[15]reportedthereason ofthisdifference,whichwasacontaminationofthegenomeofthe strainDSM30147T(GCF000421945)withpartofthegenomeofthe
strainTS43alsoisolatedbyZhangetal.[63](GCF001526605).The genomicdatawerenotincludedintheprotologueoftheemended speciesA.radiobacter,butinanycasetheemendationisnotvalid todatesinceitwasnotofficiallyperformed.
IntheworkofGanetal.[15]thegenomesofthetypestrains ofA.tumefaciensB6T(LMVK01000030.1,FCNL01000034.1)andA.
radiobacter NCPPB 3001T (LMVJ01000011.1)were obtainedand
compared withtheavailabegenomeofthestrainA.radiobacter LMG140T(MRDG01000013.1)concludingthatthetypestrainsof
A.tumefaciensandA.radiobacterrepresenttwosubspeciesfromthe samespecies.Theseauthors,takingintoaccountthatthe combina-tionAgrobacteriumradiobacterhaspriorityoverthecombination Agrobacteriumtumefaciens[49],proposedthatA.tumefaciensB6T
shouldbereclassifiedintoanewsubspeciesofA.radiobacter. How-ever,thisproposalcannotbeacceptedbecausetheprotologues ofthetwoproposedsubspecieswerenotincludedinthepaperof Ganetal.[15]andmoreoveritisnecessaryanewemendationof thespeciesA.radiobacter.Inanycase,thisproposaldoesnot mod-ifytheresolutionoftheJudicialCommissiongiventhatthespecific nameAgrobacteriumtumefacienscannotberejectedaslongasthis Commissionhasnotmadesuchdecision.
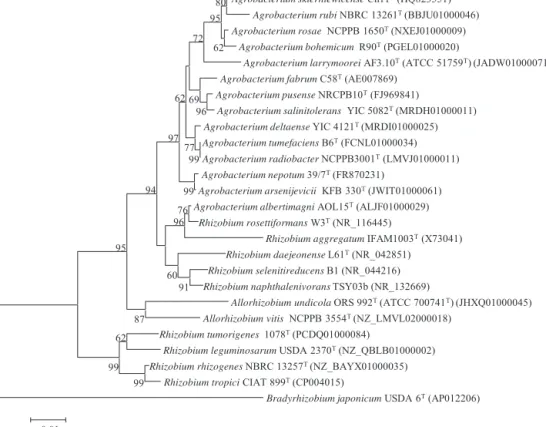
Therefore, the genus Agrobacterium currently contains 14 species if we considerthe namesA. radiobacter and A. tumefa-ciensandthespeciesA.fabrum,althoughthisspecieshasnotbeen still proposed(Table 1). One of them, A. albertimagni, is more closelyrelatedtosomeRhizobiumspeciesformingarrsgenecluster
Agrobacterium rubiNBRC 13261T(BBJU01000046) Agrobacterium rosae NCPPB 1650T(NXEJ01000009)
Agrobacterium bohemicumR90T(PGEL01000020) Agrobacterium larrymooreiAF3.10T(ATCC 51759T
Agrobacterium fabrumC58T(AE007869) Agrobacterium pusenseNRCPB10T(FJ969841)
Agrobacterium salinitoleransYIC 5082T(MRDH01000011) Agrobacterium deltaenseYIC 4121T(MRDI01000025)
Agrobacterium tumefaciensB6T(FCNL01000034)
Agrobacterium radiobacterNCPPB3001T(LMVJ01000011) Agrobacterium nepotum39/7T(FR870231)
Agrobacterium arsenijeviciiKFB 330T(JWIT01000061) Agrobacterium albertimagniAOL15T(ALJF01000029)
Rhizobium rosettiformansW3T(NR_116445)
Rhizobium aggregatumIFAM1003T(X73041)
Rhizobium daejeonenseL61T(NR_042851) Rhizobium selenitireducensB1 (NR_044216)
Rhizobium naphthalenivoransTSY03b (NR_132669)
Allorhizobium undicolaORS 992T(ATCC 700741T) (JHXQ01000045) Allorhizobiumvitis NCPPB 3554T(NZ_LMVL02000018)
Rhizobium tumorigenes1078T(PCDQ01000084)
Rhizobium leguminosarumUSDA 2370T(NZ_QBLB01000002)
Rhizobium rhizogenesNBRC 13257T(NZ_BAYX01000035) Rhizobium tropiciCIAT 899T(CP004015)
Bradyrhizobium japonicumUSDA 6T(AP012206)
99 62 99 87 95 91 76 96 60 94 96 80 62 95 99 72 69 99 77 97 62
Fig.1.Neighbour-joiningphylogeneticrootedtreebasedonrrsgenesequences(1380nt)showingthetaxonomiclocationofthespeciesfromAgrobacterium,Rhizobium andAllorhizobiumabletoinducepathogenicsymptomsinplants.Bootstrapvaluescalculatedfor1000replicationsareindicated.Bar,1ntsubstitutionper100nt.Accession numbersfromGenbankaregiveninbrackets.
(Fig.1)thatbelongtoneitherAgrobacteriumnorRhizobiumbuttoa separategenus[12,34].Nevertheless,thereclassificationofA. alber-timagniinanewgenuswillneedfurtherstudiessincecurrently thegenomesofseveralRhizobiumspeciesbelongingtothis puta-tivenewgenusarenotavailable.Theremainingspeciescurrently includedwithingenusAgrobacteriumformedarrscluster phylo-geneticallydivergenttogeneraAllorhizobiumandRhizobiumalso containingplantpathogenicspecies(Fig.1).MostofAgrobacterium speciesareabletoproducetumoursindifferentplants(Table1), neverthelessthenumberofnon-pathogenicspecieshadincresed inthelasttwoyears,withoneofthem,A.salinitolerans, ableto nodulatethelegumeSesbaniacannabina[56]andother,A.pusense, frequentlyisolatedfromclinicalsources[3],abletocausehuman sepsis[24].Ifwetakeintoaccountthatthestrainsbelongingto fivedefinedgenomospecieshavenotbeenassignedtoan Agrobac-teriumspeciesyet,asignificantincreaseinitsnumberisexpected inanearfuture.
Aknowledgements
Thisworkwassupportedby MINECO(Spanish Central Gov-ernment)Grant AGL2013-48098-P toEV. EMwasgranted by a postdoctoralcontractassociatedtothisproject.
References
[1]Allen,O.N.,Holding,A.J.(1974)GenusII.AgrobacteriumConn1942,359.In: Buchanan,R.E.,Gibbons,N.E.(Eds.),Bergey’sManualofDeterminative Bacte-riology,8thed.,Williams&Wilkins,Baltimore,pp.264–267.
[2]Arahal,D.R.,Macián,M.C.,Garay,E.,Pujalte,M.J.(2005)Thalassobius mediter-raneusgen.nov.,sp.nov.,andreclassificationofRuegeriagelatinovoransas Thalassobiusgelatinovoruscomb.nov.Int.J.Syst.Evol.Microbiol.55,2371–2376. [3]Aujoulat,F.,Marchandin,H.,Zorgniotti,I.,Masnou,A.,Jumas-Bilak,E.(2015) Rhizobium pusense is the main human pathogen in the genus Agrobac-terium/Rhizobium.Clin.Microbiol.Infect.21(472),e1–e5.
[4]Beijerinck, M.W.,van Delden, A. 1902 Über die Assimilation desfreien Stickstoffs durch Bakerien Zentralblatt für Bakteriologie, Parasitenkunde, InfektionskrankheitenundHygiene,9,AbteilungII,pp.,3–43.
[5]Bouzar,H.,Jones,J.B.(2001)Agrobacteriumlarrymooreisp.nov.,apathogen isolatedfromaerialtumoursofFicusbenjamina.Int.J.Syst.Evol.Microbiol.51, 1023–1026.
[6]Breed,R.S.,Murray,E.G.D.,Hitchens,A.P.1948Bergey’sManualof Determina-tiveBacteriology,6thed.,TheWilliams&WilkinsCo.,Baltimore,pp.,227–231. [7]Breed,R.S.,Murray,E.G.D.,Smith,N.R.1957Bergey’sManualofDeterminative
Bacteriology,7thed.,TheWilliams&WilkinsCo.,Baltimore,pp.,288–294. [8]Conn,H.J.(1938)Taxonomicrelationshipsofcertainnon-sporeformingrodsin
soil.J.Bacteriol.36,320–321.
[9]Conn,H.J.(1942)ValidityofthegenusAlcaligenes.J.Bacteriol.44,353–360. [10]Costechareyre,D.,Bertolla,F.,Nesme,X.(2009)Homologousrecombination
inAgrobacterium:potentialimplicationsforthegenomicspeciesconceptin bacteria.Mol.Biol.Evol.26,167–176.
[11]Costechareyre,D.,Rhouma,A.,Lavire,C.,Portier,P.,Chapulliot,D.,Bertolla,F., Boubaker,A.,Dessaux,Y.,Nesme,X.(2010)Rapidandefficientidentification ofAgrobacteriumspeciesbyrecAalleleanalysis:AgrobacteriumrecAdiversity. Microb.Ecol.60,862–872.
[12]deLajudie,P.,Young,J.P.W.(2018)InternationalCommitteeonSystematics ofProkaryotesSubcommitteeonthetaxonomyofrhizobiaandagrobacteria. Minutesoftheclosedmeeting,Granada,4September2017.Int.J.Syst.Evol. Microbiol.68,3363–3368.
[13]Farrand,S.K.,vanBerkum,P.B.,Oger,P.(2003)Agrobacteriumisadefinable genusofthefamilyRhizobiaceae.Int.J.Syst.Evol.Microbiol.53,1681–1687. [14]Frank,B.(1889)UeberdiePilzsymbiosederLeguminosen.Bet.Dtsch.Bot.Ges.
7,332–346.
[15]Gan,H.M.,Lee,M.V.L.,Savka,M.A.(2019)ImprovedgenomeofAgrobacterium radiobactertypestrainprovidesnewtaxonomicinsightintoAgrobacterium genomospecies4.PeerJ7,e6366.
[16]Goodner,B.,Hinkle,G.,Gattung,S.,Miller,N.,Blanchard,M.,Qurollo,B., Gold-man,B.S.,Cao,Y.,Askenazi,M.,Halling,C.,Mullin,L.,Houmiel,K.,Gordon,J., Vaudin,M.,Iartchouk,O.,Epp,A.,Liu,F.,Wollam,C.,Allinger,M.,Doughty, D.,Scott,C.,Lappas,C.,Markelz,B.,Flanagan,C.,Crowell,C.,Gurson,J.,Lomo, C.,Sear,C.,Strub,G.,Cielo,C.,Slater,S.(2001)Genomesequenceoftheplant pathogenandbiotechnologyagentAgrobacteriumtumefaciensC58.Science294, 2323–2328.
[17]Hildebrand,E.M.(1940)CanegallofbramblescausedbyPhytomonasn.sp.J. Agric.Res.61,685–696.
[18]Holmes,B.,Roberts,P.(1981)Theclassification,identificationand nomencla-tureofAgrobacteria.J.Appl.Bacteriol.50,443–467.
[19]Holt,J.G.1994Bergey’sManualofDeterminativeBacteriology,9thed.,Williams &Wilkins,Baltimore,pp.,244–254,pp.130.
[20]Keane,P.J.,Kerr,A.,New,P.B.(1970)CrowngallofstonefruitII.Identification andnomenclatureofAgrobacteriumisolates.Aust.J.Biol.Sci.23,585–596. [21]Kerr,A.,Panagopoulos,C.G.(1977)BiotypesofAgrobacteriumradiobactervar.
tumefaciensandtheirbiologicalcontrol.J.Phytopathol.90,172–179. [22]Kersters,K.,DeLey,J.(1984)GenusIII.AgrobacteriumConn1942,in:Krieg,
N.R.,Holt,J.G.(Eds.),Bergey’sManualofSystematicBacteriology,vol.1,1sted., Williams&Wilkins,Baltimore,pp.244–254.
[23]Kersters,K.,DeLey,J.,Sneath,P.H.A.,Sackin,M.(1973)Numericaltaxonomic analysisofAgrobacterium.J.Gen.Microbiol.78,227–239.
[24]Kuchibiro,T.,Hirayama,K.,Houdai,K.,Nakamura,T.,Ohnuma,K.,Tomida,J., Kawamura,Y.(2018)FirstcasereportofsepsiscausedbyRhizobiumpusensein Japan.JMMCaseRep5,e005135.
[25]Kuzmanovi ´c,N.,Puławska,J.,Proki ´c,A.,Ivanovi ´c,M.,Zlatkovi ´c,N.,Jones,J.B., Obradovi ´c,A.(2015)Agrobacteriumarsenijeviciisp.nov.,isolatedfromcrown galltumorsonraspberryandcherryplum.Syst.Appl.Microbiol.38,373–378. [26]Kuzmanovi ´c,N.,Puławska,J.,Smalla,K.,Nesme,X.(2018)Agrobacteriumrosae sp.nov.,isolatedfromgallsondifferentagriculturalcrops.Syst.Appl.Microbiol. 41,191–197.
[27]Kuzmanovi ´c,N.,Smalla,K.,Gronow,S.,Puławska,J.(2018)Rhizobium tumori-genessp.nov.,anovelplanttumorigenicbacteriumisolatedfromcanegall tumorsonthornlessblackberry.Sci.Rep.8,9051.
[28]Lapage,S.P.,Sneath,P.H.A.,Lessel,E.F.,Skerman,V.B.D.,Seeliger,H.P.R.,Clark, W.A.1992Internationalcodeofnomenclatureofbacteria,1990revision, Amer-icanSocietyforMicrobiology,Washington,D.C.
[29]Lassalle,F.,Campillo,T.,Vial,L.,Baude,J.,Costechareyre,D.,Chapulliot,D., Shams,M.,Abrouk,D.,Lavire,C.,Oger-Desfeux,C.,Hommais,F.,Guéguen,L., Daubin,V.,Muller,D.,Nesme,X.(2011)Genomicspeciesareecologicalspecies asrevealedbycomparativegenomicsinAgrobacteriumtumefaciens.Genome Biol.Evol.3,762–781.
[30]Mougel,C.,Thioulouse,J.,Perrière,G.,Nesme,X.(2002)Amathematicalmethod fordetermininggenomedivergenceandspeciesdelineationusingAFLP.Int.J. Syst.Evol.Microbiol.52,573–586.
[31]Mousavi,S.A.,Willems,A.,Nesme,X.,deLajudie,P.,Lindström,K.(2015) RevisedphylogenyofRhizobiaceae:proposalofthedelineationof Pararhizo-biumgen.nov.,and13newspeciescombinations.Syst.Appl.Microbiol.38, 84–90.
[32]Ophel,K.,Kerr,A.(1990)Agrobacteriumvitissp.nov.forstrainsofAgrobacterium biovar3fromgrapevines.Int.J.Syst.Bacteriol.40,236–241.
[33]Oren,A.,Garrity,G.M.(2016)ValidationListno.172.Listofnewnamesand newcombinationspreviouslyeffectively,butnotvalidly,published.Int.J.Syst. Evol.Microbiol.66,4299–4305.
[34]Orme ˜no-Orrillo,E.,Servín-Garcidue ˜nas,L.E.,Rogel,M.A.,González,V.,Peralta, H.,Mora,J.,Martínez-Romero,J.,Martínez-Romero,E.(2015)Taxonomyof rhi-zobiaandagrobacteriafromtheRhizobiaceaefamilyinlightofgenomics.Syst. Appl.Microbiol.38,287–291.
[35]Panday,D.,Schumann,P.,Das,S.K.(2011)Rhizobiumpusensesp.nov.,isolated fromtherhizosphereofchickpea(CicerarietinumL.).Int.J.Syst.Evol.Microbiol. 61,2632–2639.
[36]Parker,C.T.,Tindall,B.J.,Garrity,G.M.(2019)Internationalcodeof nomencla-tureofProkaryotes.Int.J.Syst.Evol.Microbiol.69,S1–S111.
[37]Peix,A.,Rivas,R.,Trujillo,M.E.,Vancanneyt,M.,Velázquez,E.,Willems,A.(2005) ReclassificationofAgrobacteriumferrugineumLMG128asHoefleamarinagen. nov.,sp.nov.Int.J.Syst.Evol.Microbiol.55,1163–1166.
[38]Puławska,J.,Willems,A.,deMeyer,S.E.,Süle,S.(2012)Rhizobiumnepotumsp. nov.isolatedfromtumorsondifferentplantspecies.Syst.Appl.Microbiol.35, 215–220.
[39]Puławska,J.,Willems,A.,Sobiczewski,P.(2012)Rhizobiumskierniewicensesp. nov.,isolatedfromtumoursonchrysanthemumandcherryplum.Int.J.Syst. Evol.Microbiol.62,895–899.
[40]Riker,A.J.,Banfield,W.M.,Wright,W.H.,Keitt,G.W.,Sagen,H.E.(1930)Studies oninfectioushairy-rootofnurseryappletrees.J.Agric.Res.41,507–540. [41]Rüger,H.J.,Höfle,M.(1992)Marinestar-shaped-aggregate-formingbacteria:
Agrobacteriumatlanticumsp.nov.;Agrobacteriummeteorisp.nov.; Agrobac-teriumferrugineumsp.nov.,nom.rev.;Agrobacteriumgelatinovorumsp.nov., nom.rev.;andAgrobacteriumstellulatumsp.nov.,nom.rev.Int.J.Syst.Bacteriol. 42,133–143.
[42]Salmassi,T.M.,Venkateswaren,K.,Satomi,M.,Newman,D.K.,Hering,J.G.(2002) OxidationofarsenitebyAgrobacteriumalbertimagni,AOL15,sp.nov.,isolated fromHotCreek,California.Geomicrobiology19,53–66.
[43]Sawada,H.,Ieki,H.,Oyaizu,H.,Matsumoto,S.(1993)Proposalforrejection ofAgrobacteriumtumefaciensandreviseddescriptionsforthegenus Agrobac-teriumandforAgrobacteriumradiobacterandAgrobacteriumrhizogenes.Int.J. Syst.Bacteriol.43,694–702.
[44]Shams,M.,Vial,L.,Chapulliot,D.,Nesme,X.,Lavire,C.(2013)Rapidandaccurate speciesandgenomicspeciesidentificationandexhaustivepopulationdiversity
assessmentofAgrobacteriumspp.usingrecA-basedPCR.Syst.Appl.Microbiol. 36,351–358.
[45]Skerman,V.B.D.,Mcgowan,V.,Sneath,P.H.A.(1980)Approvedlistsofbacterial names.Int.J.Syst.Bacteriol.30,225–420.
[46]Smith,E.F.,Townsend,C.O.(1907)Aplant-tumorofbacterialorigin.Science25, 671–673.
[47]Stapp,C.,Knosel,D.(1954)ZurGenetiksternbildenderBakterien.Zentralbl. Bakteriol.Parasitenkd.Infektionskr.Hyg.Abt.108,243–259.
[48]Starr,M.P.,Weiss,J.E.(1943)Growthofphytopathogenicbacteriainasynthetic asparaginmedium.Phytopathology33,314–318.
[49]Tindall,B.J.(2014)Agrobacteriumradiobacter(BeijerinckandvanDelden1902) Conn1942haspriorityoverAgrobacteriumtumefaciens(SmithandTownsend 1907)Conn1942whenthetwoaretreatedasmembersofthesamespecies basedontheprincipleofpriorityandRule23a,Note1asappliedtothe correspondingspecificepithets.Opinion94.JudicialCommissionofthe Inter-nationalCommitteeonSystematicsofProkaryotes.Int.J.Syst.Evol.Microbiol. 64,3590–3592.
[50]Uchino,Y.,Hamada,T.,Yokota,A.(2002)ProposalofPseudorhodobacter fer-rugineusgennov, combnov,foranon-photosynthetic marinebacterium, Agrobacteriumferrugineum,relatedtothegenusRhodobacter.J.Gen.Appl. Microbiol.48,309–319.
[51]Uchino,Y.,Hirata,A.,Yokota,A.,Sugiyama,J.(1998)Reclassificationofmarine Agrobacteriumspecies:proposalsofStappiastellulatagen.nov.,comb.nov., Stappiaaggregatasp.nov.,nom.rev.,Ruegeriaatlanticagen.nov.,comb.nov., Ruegeriagelatinovoracomb.nov.,Ruegeriaalgicolacomb.nov.,andAhrensia kieliensegen.nov.,sp.nov.,nom.rev.J.Gen.Appl.Microbiol.44,201–210. [52]ValidationListno.68,(1999)Validationofpublicationofnewnamesandnew
combinationspreviouslyeffectivelypublishedoutsidetheIJSB.Int.J.Syst.Evol. Microbiol.49,1–3.
[53]ValidationListno.92,(2003)Validationofpublicationofnewnamesandnew combinationspreviouslyeffectivelypublishedoutsidetheIJSB.Int.J.Syst.Evol. Microbiol.53,935–937.
[54]Wood,D.W.,Setubal,J.C.,Kaul,R.,Monks,D.E.,Kitajima,J.P.,Okura,V.K.,Zhou, Y.,Chen,L.,Wood,G.E.,Almeida,N.F.,Jr.,Woo,L.,Chen,Y.,Paulsen,I.T.,Eisen, J.A.,Karp,P.D.,Bovee,D.,Sr.,Chapman,P.,Clendenning,J.,Deatherage,G.,Gillet, W.,Grant,C.,Kutyavin,T.,Levy,R.,Li,M.J.,McClelland,E.,Palmieri,A.,Raymond, C.,Rouse,G.,Saenphimmachak,C.,Wu,Z.,Romero,P.,Gordon,D.,Zhang,S.,Yoo, H.,Tao,Y.,Biddle,P.,Jung,M.,Krespan,W.,Perry,M.,Gordon-Kamm,B.,Liao, L.,Kim,S.,Hendrick,C.,Zhao,Z.Y.,Dolan,M.,Chumley,F.,Tingey,S.V.,Tomb, J.F.,Gordon,M.P.,Olson,M.V.,Nester,E.W.(2001)Thegenomeofthenatural geneticengineerAgrobacteriumtumefaciensC58.Science294,2317–2323. [55]Yan,J.,Li,Y.,Han,X.Z.,Chen,W.F.,Zou,W.X.,Xie,Z.,Li,M.(2017)Agrobacterium
deltaensesp.nov.,anendophyticbacteriaisolatedfromnoduleofSesbania cannabina.Arch.Microbiol.199,1003–1009.
[56]Yan,J.,Li,Y.,Yan,H.,Chen,W.F.,Zhang,X.,Wang,E.T.,Han,X.Z.,Xie,Z.H.(2017) Agrobacteriumsalinitoleranssp.nov.,asaline-alkaline-tolerantbacterium iso-latedfromrootnoduleofSesbaniacannabina.Int.J.Syst.Evol.Microbiol.67, 1906–1911.
[57]Young,J.M.(2004)RenamingofAgrobacteriumlarrymooreiBouzarandJones 2001asRhizobiumlarrymoorei(BouzarandJones2001)comb.nov.Int.J.Syst. Evol.Microbiol.54,149.
[58]Young,J.M.(2008)Agrobacterium-taxonomyofplant-pathogenicRhizobium species.In:Tzfira,T.,Citovsky, V.(Eds.),Agrobacterium:FromBiologyto Biotechnology,Springer,NewYork,NY,pp.183–220.
[59]Young,J.M.,Kerr,A.,Sawada,H.(2005)Agrobacterium,in:Brenner,D.J.,Krieg, N.R.,Staley,J.T.,Garrity,G.M.(Eds.),TheAlpha-,Beta-,Delta-and Epsilon-proteobacteria,TheProteobacteria,PartC,Bergey’s Manualof Systematic Bacteriology,vol.2,2nd.Ed.,Springer,NewYork,pp.340–345.
[60]Young,J.M.,Kuykendall,L.D.,Martínez-Romero,E.,Kerr,A.,Sawada,H.(2001) ArevisionofRhizobiumFrank1889,withanemendeddescriptionofthegenus, andtheinclusionofallspeciesofAgrobacteriumConn1942andAllorhizobium undicoladeLajudieetal.1998asnewcombinations:Rhizobiumradiobacter,R. rhizogenes,R.rubi,R.undicolaandR.vitis.Int.J.Syst.Evol.Microbiol.51,89–103. [61]Young,J.M.,Kuykendall,L.D.,Martínez-Romero,E.,Kerr,A.,Sawada,H.(2003) ClassificationandnomenclatureofAgrobacteriumandRhizobium—areplyto Farrandetal.(2003).Int.J.Syst.Evol.Microbiol.53,1689–1695.
[62]Zahradník,J.,Nunvar,J.,Paˇrízková,H.,Koláˇrová,L.,Palyzová,A.,Mareˇsová,H., Grulich,M.,Kyslíková,E.,Kyslík,P.(2018)Agrobacteriumbohemicumsp.nov. isolatedfrompoppyseedwastesincentralBohemia.Syst.Appl.Microbiol.41, 184–190.
[63]Zhang,L.,Li,X.,Zhang,F.,Wang,G.(2014)GenomicanalysisofAgrobacterium radiobacterDSM30147TandemendeddescriptionofA.radiobacter(Beijerinck
andvanDelden1902)Conn1942(ApprovedLists1980)emend.Sawadaetal. 1993.Stand.GenomicSci.9,3.