www .e l s e v i e r . c o m / l o c a t e / b j i d
The
Brazilian
Journal
of
INFECTIOUS
DISEASES
Original
article
Detection
of
E.
coli
O157:H7
and
Shigella
dysenteriae
toxins
in
clinical
samples
by
PCR-ELISA
Jafar
Amani
a,
Askary
Ahmadpour
a,
Abbas
Ali
Imani
Fooladi
a,∗,
Shahram
Nazarian
baAppliedMicrobiologyResearchCenter,BaqiyatallahUniversityofMedicalSciences,Tehran,Iran
bImamHossainUniversity,FacultyofScience,DepartmentofBiology,Tehran,Iran
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received14October2014 Accepted21February2015 Availableonline21April2015
Keywords:
Shigelladysenteriae
E.coliO157:H7
PCR-ELISA
Diagnosticmethods
a
b
s
t
r
a
c
t
Shigatoxinproducing bacteriaare potentialcauses ofserioushumandisease suchas hemorrhagiccolitis,severeinflammationsofileocolonicregionsofgastrointestinaltract, thrombocytopenia,septicemia, malignantdisorders inurinaryducts, hemolyticuremic syndrome(HUS).Shiga toxin1(stx1),shigatoxin2(stx2),ora combinationofbothare responsibleformostclinicalsymptomsofthesediseases.Alotofmethodshavebeen devel-opedsofartodetectshigatoxinssuchascellculture,ELISA,andRFPLA,butduetohighcosts andlabortimeinadditiontolowsensitivity,theyhavenotreceivedmuchattention.Inthis study,PCR-ELISAmethodwasusedtodetectgenesencodingshigatoxins1and2(stx1and stx2).Todetectstx1andstx2genes,twoprimerpairsweredesignedforMultiplex-PCRthen PCR-ELISA.PCRproducts(490and275,respectively)weresubsequentlyverifiedby sequenc-ing.SensitivityandspecificityofPCR-ELISAmethodweredeterminedbyusinggenome serialdilutionandEnterobacteriastrains.PCR-ELISAmethodusedinthisstudyprovedtobe arapidandpreciseapproachtodetectdifferenttypesofshigatoxinsandcanbeusedto detectbacterialgenesencodingshigatoxins.
©2015ElsevierEditoraLtda.Allrightsreserved.
Introduction
Shigatoxinsbelongtoalargefamilyofbacterialtoxinswith two major groups, stx1 and stx2.1 These virulence factors aremainlyproducedbyShigelladysenteriaeandShigatoxigenic group ofEscherichia colilike E. coli O157:H7 which are able tocauseinfectious diseases.2,3 ThetoxinisoneoftheAB
5
toxins and hasbinding (B) and catalytic domains (A). The pentamericBsubunitofthetoxinisresponsibleforreceptor
∗ Correspondingauthorat:AppliedMicrobiologyResearchCenter,BaqiyatallahUniversityofMedicalSciences,VanakSq.MolasadraSt.,
P.O.Box19395-5487,Tehran,Iran.
E-mailaddress:imanifouladi.a@gmail.com(A.A.ImaniFooladi).
bindingandintracellulartraffickingoftheholotoxins.4The toxinbindstoGb3locatedoncellsurfacesandisintroduced by endocytic uptake. N-glycosidase activity of the A sub-unit inhibits protein synthesis in the cell and causes cell death.5Insomecellsthesetoxinsalsotriggercytokine syn-thesis and induce apoptosis, which is caused by ribotoxic stress.6 The gene encoding toxin in Shigella dysenteriae is chromosomal.HoweverstxgeneinE.coliO157:H7is associ-atedwithaprophage.7Differentsubtypesofshigatoxinare identifiedasstx1,stx1c,stxfc,stx2,stx2e,stx2dandstx2g.8
http://dx.doi.org/10.1016/j.bjid.2015.02.008
Infections by shiga toxins producing bacteria have world-wideprevalenceandarewidespreadindevelopingcountries suchassouth-easternAsiancountries,Indiansubcontinent, SouthAfrica,centralAsia,andBangladesh.9InfectionofShiga toxin producing bacteria is a major health concern even in developed countries all over the world. These bacteria are potentialcause ofdiarrhea,hemorrhagic colitis,severe inflammationsofileocolonic regionsofthegastrointestinal tract,thrombocytopenia,septicemia,centralnervoussystem (CNS)involvement,malignantdisordersinurinaryducts,and hemolyticuremic syndrome(HUS).10 urinarytractinfection byEHECismainlygeneratedbyE.coliO157.Inchildrenunder fiveyearsoldandadultsover60,astheyhavethereceptor, itcauseskidneyfunctiondeficiencyandhasadeathrateof 5–10%.11 Cows,goatsand other animalscannaturally bea sourceofstxproducingE.coliandotheranimalssuchascrabs alsoplayarole initstransfer.12 Transferbetweenhumans canalsotakeplace.13Sinceshigatoxinscausemanydiseases, especiallyinchildrenandimmunocompromisedelderly peo-ple,arapidandsensitivediagnosticmethodwithprognostic informationwould berather useful.So far,many different detectionmethodssuchascellculture,serologicaland molec-ularmethodssuchasRPLA,real-time,PCR,hybridizationhave beenutilizedtodetectshigatoxinsortheirrespectivegenes.14 Yet,allthesemethodshavetheirownshortcomingsasthey are time-consuming, quite costly and have limitations in handlingmanysamplessimultaneously.Atpresent,although molecularmethodssuchas PCRandhybridization, despite beingless time-consuming,less costly,andmoresensitive, theyare notsuitable astheyrelyon agarose electrophore-siswithcarcinogenethidiumbromide,amajorhealththreat forlabpersonnel,anddoesnotallowanalysisofmany sam-plesatatimeincaseofepidemicbreakouts.15Nevertheless, real-timePCRdespite100%specificityandhighsensitivityhas notgainedmuchattentionbecauseofhighcostsof fluores-centmaterialandshortageofexpertpersonnel.16Toovercome shortcomingsoftheaforementionedmethods,PCR-ELISAis anappropriatealternativeapproachfordetectingstxgenes, whichissafeandnon-radioactive.PCR-ELISAismore conve-nientforrapidand reliabledetection and quantificationof pathogen-specificgene sequences.17,18 Besideshavingbeen usedinmedicalandfoodindustries,PCR-ELISAhasalsobeen usedintheveterinaryindustry.19
Inthis study,specific primersand probes forstx genes, amplificationandlabelingofproductsDIG-dUTP, hybridiza-tion ofstreptavidinwith biotinylatedprobes and detection with antibody against conjugated digoxigenin and peroxi-daseinmicrotiterplatesweredesigned.Weidentifiedspecific sequencesofstxgenesinalargenumberofsamplesusing smallamountssimultaneouslywithconsiderablesensitivity andshortturnaroundtime.
Materials
and
methods
Bacterialstrains
ShigelladysenteriaeandE.coliO157:H7strainswereprovided
fromShahedUniversity,Tehran,IranandPseudomonas
aeru-ginosa, Salmonellatyphimurium,Salmonellaparatiphi, Klebsiella
pneumoniaandVibriocholerastrainswereprovidedby
Baqiy-atallah University of Medical Sciences, Tehran, Iran. The strainswereverifiedbybiochemicalandimmunologic meth-ods.
ExtractionofbacterialgenomicDNA
ToextractgenomicDNAfrom ShigelladysenteriaeandE.coli O157:H7bacteriawereincubatedinliquidLBmediumfor18h in 37◦C.Bacterial culture was centrifuged in 3000rpmfor
5min.Thepelletwasresuspendedin300LTEbufferfollowed bylysissolutioncontaining10Llysozyme(10mg/mL),200L SDS20%,3LproteinaseKandincubatedin37◦Cfor60min.
DNAwaspurifiedbyextractionwithanequalvolumeof phe-nol/chloroform/isoamylalcohol(25:24:1)inthepresenceof5M sodiumperchlorate.A1/10volumeof3Msodiumacetateand 2volumesofabsoluteethanolwereaddedandincubatedin −20◦Cfor13h.Thegenomewaspelletedbycentrifugation, washed with70% ethanol and dried.Finally, DNA samples weredissolvedin100LTEbufferand,toeliminateRNA,3L RNaseAwasaddedandthetubeswereincubatedin37◦Cfor
30min.
Theconcentration andpurityofthe DNAsampleswere determinedspectrophotometricallyatA260andA280by Nan-oDrop2000,ThermoScientific(USA).
Primerandprobedesign
Todesignprimers andthe nucleic-acid sequencingprobes, centralpartsofshigatoxingeneswereusedastemplate. Fea-turesinthedesignedprimerssuchasGCcontent;Tm,G,etc. were checkedbyDNASISand Oligo7softwares.Theprimer andprobesequencesareillustratedinTable1.The oligonu-cleotidesweresuppliedbyCinnaClone(IRAN).
Isolationandamplificationofstx1andstx2genesbyPCR
PCRreagentsinafinalvolumeof25Lincluded: 1L tem-plateDNA,0.5LTaqDNApolymerase(5U/L),1Lofeach primer(10pmol/L),1LdNTPmixture,2.5L10×PCRbuffer, 1.5LMgCl2(50mM)and16.5LsterileDDW.Thermalcycling
ofamplificationmixturewasperformedin30cycles.20 ThePCRprogramwas carriedout at94◦Cfor3min
fol-loweddenaturingfor45sat94◦C,annealingfor45sat59◦C
and an extension for 1min at 72◦C. The final extension
was at72◦C for5min. PCRproductswere electrophoresed
in 1%agarosefollowed bystainingwithethidium bromide
(0.5g/mL) then visualized under ultraviolet light, and the
resultswererecordedbyphotography.PCRproductswere ver-ifiedbysequencing.ForlabelingofPCRproducts,thereaction ofPCRwas performedbydNTP mixturecontaining digoxi-geninlabelingmix(DigoxigenindNTPs,Roche,Germany)with thesamecondition.
DetectionofPCRproductsbyELISA
Table1–PCRprimersandhybridizationcaptureprobeforstx1andstx2genes.
Oligonucleotide Sequence Nucleotideposition Expectproductsize(bp)
Ahstx1F TTGTTTGCAGTTGATGTCAGAGG 210–233 275
Ahstx1R CAGGCAGGACACTACTCAACCTTC 676–700
Ahstx2F TTGCTGTGGATATACGAGGGC 214–235 490
Ahstx2R CGCCAGATATGATGAAACCAGTG 466–489
pStx1 Biotin-TGTTGCAGGGATCAGTCGTACG 422–444 –
pStx2 Biotin-CATATATCAGTGCCCGGTGTG 355–376 –
productofstx1 andstx2geneswere addedto90L1×SSC
bufferin1.5mLtubesandincubatedinboilingwaterfor10min andthen5minonice.Inthenextstep,10Lofstx1andstx2
probeswereaddedtoeachtubeandafter2hin60◦C,100L
fromthishybridizationsolutionwereaddedtoeachwelland after1hin37◦C,itwaswashedwith20%BPSTbuffer3times.A
1/1000solutionofanti-digoxigeninantibodyconjugatedwith peroxidesinPBSTbufferwaspreparedand100Lofthis
solu-tionwasaddedtoeachwell(includingcontrols)andafter1h in37◦Ctheplateswerewashedanddriedasdescribed
ear-lier.100Lofsubstratesolution(2mgOPD,100Ldetection
buffer,5L30%hydrogenperoxide)wasaddedtowells.100L
of1MH2SO4wasaddedtostopthereaction.21Theoptical den-sitywasmeasuredat490nmusinganELISA reader(Dynex Technologies,Guornesey,ChannelIslandsandGreatBritain).
EvaluationPCRspecificityandsensitivity
Todeterminethespecificity,PCRwascarriedoutwithgenomic DNA extracted from Pseudomonas aeruginosa, Salmonella
typhimurium, Salmonella paratiphiC, Klebsiellapneumonia and
Vibriocholera.Productswereanalyzedon1%agarosegel.To
determine the minimum genomic DNA concentration that couldbedetectedbythemethod,seriallydilutedgenomicDNA inTEbuffer(pH=8)wasusedasPCRtemplateandtheproduct wasanalyzedon1%agarosegel.
SensitivityevaluationofPCR-ELISAtechniqueusing labeledPCRproductsofstx1andstx2
To determine the detection limit for stx gene, genomic DNA was extracted and tenfold serial dilutions containing 108ng/Lto0.108pg/Land156ng/Lto0.156pg/LofE.coli O157:H7and Shigelladysenteriae genomicDNA,respectively, werepreparedandallthestepswerecarriedoutasdescribed previously.22
PCR-ELISAspecificityevaluationusingbacterialstrains
ToevaluatespecificityofPCR-ELISA,E.coliO157:H7,Shigella
dysenteriae, Pseudomonas aeruginosa, Salmonella typhimurium,
Salmonella paratiphiC and Klebsiellapneumonia strains were
growninLBmedium.GenomicDNAswereextractedandafter evaluatingtheirconcentrationbyNanoDrop,10−2folddilution
waspreparedastemplates.ThePCRwascarriedout accord-ingtomentionedprotocolsandtheproductswereanalyzed byagarosegelelectrophoresisandELISA.22
Clinicalsamplesanalysis
Inthisstudy63positivesamplesofShigelladysenteriaeand E.coliO157:H7obtainedfromstoolandurinecultureswere analyzed.SamplesweregatheredfromMazandaranhospitals ina3-monthperiodbetweenJanuaryandMarchof2013.Age groupsincludedchildrenbetween8monthsto10yearsold, adultsbetween20and30 andbetween50and 90yearsold (Table2).Samplesweretransferredtothelab,cultured,and aftergrowthwereidentifiedindifferentialandspecificmedia andverifiedbyspecificantiserums.FollowingDNAextraction byboilingmethod,PCR-ELISAwascarriedout.Dataanalysis wasperformedonSPSS.
Results
PCR
DNA from extracted genome of bacteria cultured in LB mediumwas availableinlargequantitiesand was ofgood quality. Thepurity ofthe DNA sampleswas confirmed by absorbance(A260/A280)ratio,whichwas1.8–2.0.PCRwas per-formedforeachstrainwithspecificprimers.EachPCRproduct wasobtainedasclearbandat275and490bpgeneratedbystx2 andstx1genesrespectively(Fig.1b).ThesizesofPCRproducts werethesameaspredicted.
1 2 3 2 1
490bp
275bp
500bp
300bp
a
b
Fig.1–AgarosegelelectrophoresisofextractedE.coli
O157:H7andShigelladysenteriaegenomicDNAandanalysis
Table2–Clinicalcharacteristicsofpatients.
PatientID Age(years) Sex Typeofspecimen Clinicalmanifestations Stxgenotype
Diarrhea Abdominalpain
2 30 F Bloodystool + + Stx1/Stx2
3 20 F Bloodystool + + Stx1/Stx2
8 8 M Bloodystool + + Stx1/Stx2
10 6 M Bloodystool + + Stx2
14 20 F Bloodystool + − Stx2
15 56 M Bloodystool + − Stx2
18 20 M Bloodystool + − Stx2
20 6 F Bloodystool + + Stx2
23 9 M Waterystool + − Stx2
27 8 M Bloodystool + + Stx1/Stx2
32 57 M Bloodystool + − Stx2
35 9 M Bloodystool + − Stx2
43 25 F Bloodystool + + Stx2
47 50 F Bloodystool + − Stx2
53 7 M Bloodystool + + Stx2
55 55 F Waterystool + − Stx2
59 8 M Bloodystool + − Stx2
a
b
275bp 490bp
7 6 5 4 3 2 1 1 2 3 4 5 6 7
Fig.2–SpecificityofPCRdetectionofE.coliO157:H7(a)andShigelladysenteriae(b).Lane1,positivecontrol;Lane2,100bp DNAladder;Lane3,Pseudomonasaeruginosa;Lane4,Klebsiellapneumonia;Lane5,Vibriocholera;Lane6,Salmonella typhimurium;Lane7,SalmonellaparatyphiC.
SpecificityofPCR
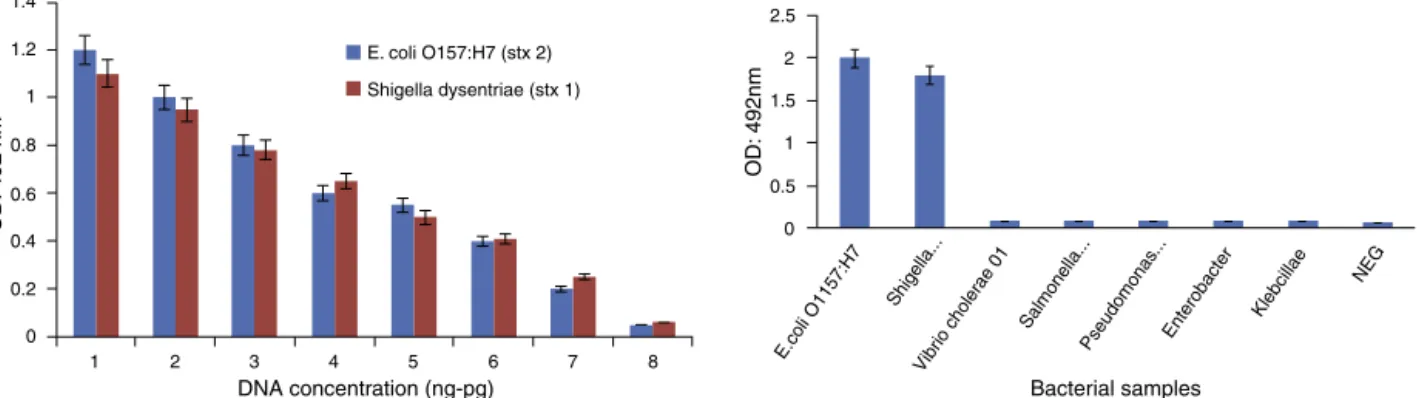
AftervalidationofPCRproducts,specificity ofthereaction was examined using genomes of Pseudomonas aeruginosa,
Salmonellatyphimurium,SalmonellaparatiphiC,Klebsiella pneu-moniaandVibriocholera.AsshowninFig.2,nocross-reaction
was found between the individual primers and non-target pathogensinthePCR.
SensitivityofPCR
After preparing a serial dilution from E. coli O157:H7 and
Shigella dysenteriae ata primary concentration of108ng/L
1 2 3 4 5 6 7 8 1 2 3 4 5 6 7 8
a
b
275bp
490bp
490bp
275bp
1 2 3 4 5
Fig.4–RepresentativeelectrophoreticgelsofPCRproducts labeledwithdigoxigenin.Lane1,100bpDNAladder;Lane 2,PCRproductofstx1fromShigelladysenteriaegenomic DNA;Lane3,PCRproductofstx1fromShigelladysenteriae
genomicDNAlabelingwithdNTPDIGmix;Lane4,PCR
productofstx2fromE.coliO157:H7genomicDNA;Lane5,
PCRproductofstx2fromE.coliO157:H7genomicDNA
labelingwithdNTPDIGmix.
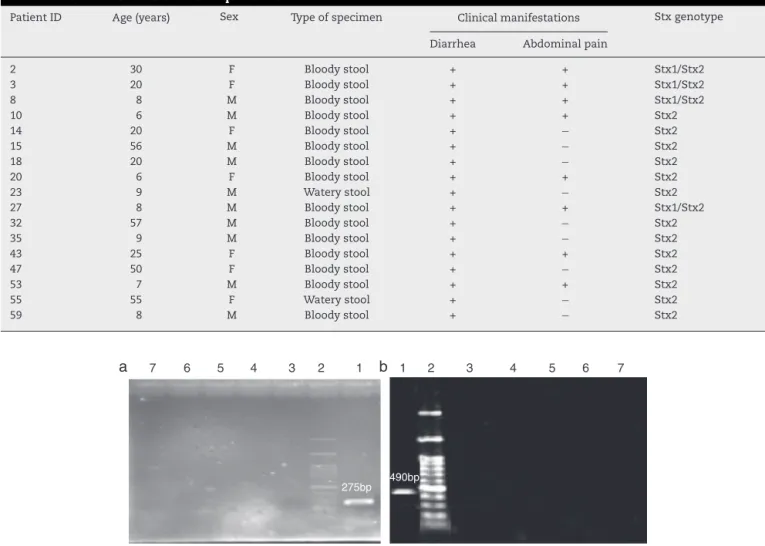
and 156ng/L,the PCRreactions were performedon these dilutionsandtheresultsaredisplayedinFig.3.Regarding con-centrationsoftheprimarysample(108ng/Land156ng/L), sensitivityofthereactionwas calculatedas1.08pg/Land 1.56pg/L,respectively.
SpecificPCRwithdNTPDIGlabelingmix
APCRreactionwasperformedwithdigoxigeninlabelingmix andtheresultswereanalyzedon1%agarosegel(Fig.4).
PCR-ELISAspecificityandsensitivityassay
ThespecificityofthePCR-ELISAwasanalyzedusinggenomes
ofPseudomonasaeruginosa,Salmonellatyphimurium,Salmonella
paratiphiC,KlebsiellapneumoniaandVibriocholera.
Todetermine the minimumdetectable concentration of genomic DNA of Shigella dysenteriae and E. coli O157:H7, serialdilutionswere subjectedtothePCR-ELISAtechnique. Theresults(Fig. 5)demonstratethe possibilityofdetecting 1.08pg/Land1.56pg/LE.coliO157:H7andShigella dysente-riae,respectively.
Clinicalresults
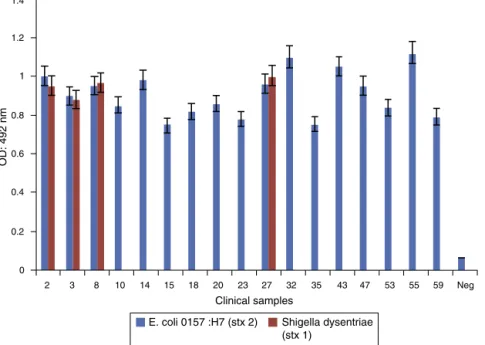
Atotalof63clinicalsampleswerecollectedandscreenedfor thepresenceofE.coliO157:H7andShigelladysenteriaestrains.
Seventeen(26.98%)samplesweredetectedasstxpositive. Agedistributionofthepatientsrangedfrom<1to90years. E.coliO157:H7andShigelladysenteriaeinfectedpatientshad an average age of 23.17 years, and 47.05% were less than 10 years old (Table 2). Themost common symptoms were bloodystools(88.23%)andabdominalpain(47.05%).Sex dis-tribution ofE. coli O157:H7 and Shigella dysenteriaeinfected patientswere7females(41.18%),and10males(58.82%). Bac-teriologicalcultureresultswerecomparedtothoseobtained byPCR-ELISA.ResultsobtainedfromPCR-ELISAassay(Fig.6) andfromselectiveculturewerecompared.Therewasa sig-nificantassociationbetweenPCR-ELISAandcultureresultsfor detectingandscreeningE.coliO157:H7andShigelladysenteriae. (p<0.001).
Discussion
Shigatoxinproducingbacteriainfectionisamajorhealth con-cernevenindevelopedcountriesallovertheworld.9,23,24With increasingreports ofE.coliO157:H7 and Shigelladysenteriae infections,greatattentionhasbeengiventothedevelopment ofmethodsfordetectingthesepathogensandapproachesfor preventionandtreatmentoftheseinfections.Thisstudywas intendedtoreachpartoftheseaims.
Inrecentyears,ELISAtesthasbeendesignedtodetectshiga toxinsdirectlyinstoolsamples.Thetestisrapidandhasa goodpotentialforshigatoxindetectionsinceitcandetectthe presenceofshigatoxin-producingE.coli(STEC)orothershiga toxin-producingbacteria.Sinceshigatoxintype differentia-tionrequireshighcostmonoclonalandpolyclonalantibodies it isnotwidelyused.Hybridization methodisaneffective, highly sensitive and specificmolecular method forprecise detectionofshigatoxins,andusesnon-radioactivesubstances suchasdigoxigeninandbiotin.However,sinceitisnot suit-ableforusinginlargenumberofclinicalsamples,thismethod isnotusedinmanyclinicallabs.25
In contrast toserological and microbiologicaltests, PCR providesarapidandsensitivealternative.Thistechnique,first developedbyKarchandMeyer,includesaprimerpairfroma
1.4
1.2
1
0.8
0.6
0.4
0.2
0
1 2 3 4 5 6 7 8
DNA concentration (ng-pg)
E. coli O157:H7 (stx 2)
Shigella dysentriae (stx 1)
0.5 1.5
1
0 2 2.5
OD:
492nm
Bacterial samples
E.coli O1157:H7 Shigella...
NEG
Vibr io cho
lerae 01 Salmonella...
Pseudomonas ...
Enterobacter Klebcillae
OD:
492 nm
0
2 3 8 10 14 15 18 20 23 27 32 35 43 47 53 55 59 Neg
Clinical samples
0.2 0.4 0.6 0.8 1 1.2 1.4
OD: 492 nm
E. coli 0157 :H7 (stx 2) Shigella dysentriae (stx 1)
Fig.6–PCR-ELISAanalysisofclinicalsamples,E.coliO157:H7(stx2)andShigelladysenteriae.(stx1).Negativecontrolsare alsoindicated.Assaycut-offwascalculatedbyreplicateanalysisofnegativesamples(OD:0.125).
conservedregionofstx1andstx2inhomologousgeneswhose maindefectswere lowTm and ineffectivenessindifferent typesofshigatoxins.15Inthatregard,todetectdifferenttypes ofshigatoxinsitisnecessarytodesignamultiplexPCRwithat leasttwopairsofprimerstodetectshigatoxingene.Thefirst studyonmultiplexPCRdetectionshigatoxinswasbyCebula etal.whodesignedspecificprimersandusedafragmentas positiveinternalcontrol.Nonetheless,primerswereonlyable todetectstx2.26SubsequentstudieswerecarriedoutbyPaton etal.,Passetal.,Philotetal.,andBelangeretal.Theirmain dis-advantagewasthatallfragmentswerelow-sizeandwerehard todissolveinagarosegel.14,27,28Theprimersthatwedesigned forthisstudy,aftercomparisonwithavailabledatafromgene banks,using softwareand experimenton clinicalsamples, provedtolacktheabovementioneddisadvantagesandtobe ableofamplifydifferentshigatoxingenes.Inaddition,these amplifiedfragmentswereeasilydissolvedinagarosegeland bothstx1andstx2genescouldbesimultaneouslydetectedin clinicalsamples.OnepotentialproblemofPCRisthepresence ofinhibitors,whichmaycausefalseresultsofthetest.InGram negativebacteriadifferentlipids,carbohydrates,andproteins presentincellwallcanactasinhibitorsandreducethe sensi-tivityofthereaction.29Toovercometheselimitationsandto increasesensitivity,bacterialgenomemustbepurifiedbefore carryingoutthereaction.Franketal.in1998usedthe boil-ingmethodtoextractgenomicDNA;however,inthismethod inhibitorscouldnotbeeliminated.30Wangetal.in2002used SDS,lysozyme,tris,glucoseandEDTAforextractionwhich increasedsensitivityto100pmol/L.31In2011,Marzonyetal. removedinhibitorsandextractedgenomicDNAbyusingCTAB and5MNaClandreportedasensitivityof2.1pg/L.32 Com-paringPCR-ELISAwithVerocellassays,Fachetal.33showed thatresultswithPCRwereobtainedwithin24hwhereaswith Vero cell assaysresultswere available onlyafterfivedays. Furthermore,Beilei et al.developedPCR-ELISA methods to
detectEscherichiacoliO157:H7andotherShigatoxin-producing E. coli (STEC)in food, where105 CFU ofSTEC per gram of
groundbeefdetectedwithoutanycultureenrichmentby PCR-ELISA.34
In this study, inhibitors were effectively eliminated by using TE buffer, lysozyme, 20% SDS, proteinase K, RNase A,3MSodiumAcetateand coldisopropanol.Consequently, sensitivity of the test improved to 1.08pg/L. PCR-ELISA was successfully carried out with genomic DNA extracted fromclinicalsamples;fromShigelladysenteriae,a490bp frag-mentwasamplifiedasmentionedearlier.ForE.coliO157:H7 both490and 275bpfragmentswereamplified demonstrat-ing that it can produce both toxins. PCR-ELISA correctly confirmed the specific stx1 and stx 2 genes. Non-positive results for other strains indicate that the tests are highly specific. Moreover, we have compared the performanceof PCR-ELISA with standard agarose gel electrophoresis. PCR-ELISAassaywasmoresensitivethanthegelelectrophoresis. Our data indicate that PCR-ELISA is highly specific, and hashighersensitivitythanconventionalgelelectrophoresis. By offering shorter turnaround time and high sensitiv-ity, PCR-ELISA has the potential to serve as a powerful detectiontoolinmedicineandinfoodandagricultural indus-tries.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgement
submittedto AppliedMicrobiology Research Center, Baqiy-atallahUniversityofMedicalSciences,Tehran,Iraninpartial fulfillmentoftherequirementsfortheMScinmicrobiology.
r
e
f
e
r
e
n
c
e
s
1. PatonJC,PatonAW.PathogenesisanddiagnosisofShiga toxin-producingEscherichiacoliinfections.ClinMicrobiolRev. 1998;11:450–79.
2. KarmaliMA.InfectionbyShigatoxin-producing
Escherichiacoli.MolBiotechnol.2004;26:117–22.
3. KarmaliMA.Prospectsforpreventingserioussystemic toxemiccomplicationsofshigatoxin-producingEscherichia coliinfectionsusingshigatoxinreceptoranalogues.JInfect Dis.2004;189:355–9.
4. LingwoodCA.Shigatoxinreceptorglycolipidbinding. MethodsMolMed.2002;73:165–86.
5. EndoY,TsurugiK,YutsudoT,TakedaY,OgasawaraT,Igarashi K.SiteofactionofaVerotoxin(VT2)fromEscherichiacoli
O157:H7andofShigatoxinoneukaryoticribosomes.EurJ Biochem.1988;171:45–50.
6. FosterGH,TeshVL.Shigatoxin1-inducedactivationofc-Jun NH2-terminalkinaseandp38inthehumanmonocyticcell lineTHP-1:possibleinvolvementintheproductionofTNF-␣.J
LeukocBiol.2002;71:107–14.
7. HuangA,FriesenJ,BruntonJ.Characterizationofa bacteriophagethatcarriesthegenesforproductionof Shiga-liketoxin1inEscherichiacoli.JBacteriol. 1987;169:4308–12.
8. GobiusKS,HiggsGM,DesmarchelierPM.Presenceof activatableShigatoxingenotype(stx2d)inShigatoxigenic
Escherichiacolifromlivestocksources.JClinMicrobiol. 2003;41:3777–83.
9. SackD,HoqueS,EtheridgeM,HuqA.Isprotectionagainst shigellosisinducedbynaturalinfectionwithPlesiomonas shigelloides?Lancet.1994;343:1413–5.
10.SieglerRL.Spectrumofextrarenalinvolvementin postdiarrhealhemolytic-uremicsyndrome.JPediatrics. 1994;125:511–8.
11.SieglerRL.PostdiarrhealShigatoxin-mediatedhemolytic uremicsyndrome.JAMA.2003;290:1379–81.
12.UrdahlA,BeutinL,SkjerveE,ZimmermannS,WastesonY. AnimalhostassociateddifferencesinShigatoxin-producing
Escherichiacoliisolatedfromsheepandcattleonthesame farm.JApplMicrobiol.2003;95:92–101.
13.KumarH,KarunasagarI,KarunasagarI,TeizouT,ShimaK, YamasakiS.CharacterisationofShigatoxin-producing
Escherichiacoli(STEC)isolatedfromseafoodandbeef.FEMS MicrobiolLett.2004;233:173–8.
14.PhilpottD,EbelF.E.coli:shigatoxinmethodsandprotocols. Springer;2003.
15.KarchH,MeyerT.Singleprimerpairforamplifyingsegments ofdistinctShiga-like-toxingenesbypolymerasechain reaction.JClinMicrobiol.1989;27:2751–7.
16.CroxenMA,FinlayBB.Molecularmechanismsof
Escherichiacolipathogenicity.NatRevMicrobiol.2010;8:26–38.
17.DalyP,CollierT,DoyleS.PCR-ELISAdetectionof
Escherichiacoliinmilk.LettApplMicrobiol.2002;34:222–6.
18.PerelleS,DilasserF,MalornyB,GroutJ,HoorfarJ,FachP. ComparisonofPCR-ELISAandLightCyclerreal-timePCR assaysfordetectingSalmonellaspp.inmilkandmeat samples.MolCellProbes.2004;18:409–20.
19.KobetsT,BadalováJ,GrekovI,HavelkováH,SvobodováM, LipoldováM.Leishmaniaparasitedetectionand
quantificationusingPCR-ELISA.NatProtoc.2010;5:1074–80.
20.AhmadpourA,AmaniJ,ImaniFooladiAA,SedighianH, NazarianS.DetectionofShigelladysenteriaeandE.coliO157:H7 toxinsbymultiplexPCRmethodinclinicalsamples.IranJ MedMicrobiol.2013;7:41–51.
21.MousaviSL,NazarianS,AmaniJ,RahgerdKarimiA.Rapid screeningoftoxigenicvibriocholeraeO1strainsfromsouth IranbyPCR-ELISA.IranBiomedJ.2008;12:15–21.
22.MousaviSL,SalimiyanJ,RahgerdiAK,AmaniJ,NazarianS, ArdestaniH.ArapidandspecificPCR-ELISAfordetecting
Salmonellatyphi.IranJClinInfectDis.2006;1:113–9.
23.SandvigK.Shigatoxins.Toxicon.2001;39:1629–35.
24.KargarM,DaneshvarM,HomayounM.Surveillanceof virulencemarkersandantibioticresistanceofshigatoxin producingE.coliO157:H7StrainsfromMeatsPurchasein Shiraz.ISMJ.2011;14:76–83.
25.JacksonMP.DetectionofShigatoxin-producingShigella dysenteriaetype1andEscherichiacolibyusingpolymerase chainreactionwithincorporationofdigoxigenin-11-dUTP.J ClinMicrobiol.1991;29:1910–4.
26.CebulaTA,PayneWL,FengP.Simultaneousidentificationof strainsofEscherichiacoliserotypeO157:H7andtheirShiga-like toxintypebymismatchamplificationmutation
assay-multiplexPCR.JClinMicrobiol.1995;33:248–50.
27.BélangerSD,BoissinotM,MénardC,PicardFJ,BergeronMG. RapiddetectionofShigatoxin-producingbacteriainfecesby multiplexPCRwithmolecularbeaconsonthesmartcycler.J ClinMicrobiol.2002;40:1436–40.
28.WongCS,JelacicS,HabeebRL,WatkinsSL,TarrPI.Theriskof thehemolytic–uremicsyndromeafterantibiotictreatmentof
EscherichiacoliO157:H7infections.NEnglJMed. 2000;342:1930–6.
29.TamminenM,JoutsjokiT,SjöblomM,etal.Screeningoflactic acidbacteriafromfermentedvegetablesbycarbohydrate profilingandPCR-ELISA.LettApplMicrobiol.2004;39:439–44.
30.FranckSM,BosworthBT,MoonHW.MultiplexPCRfor enterotoxigenic,attachingandeffacing,andShiga toxin-producingEscherichiacolistrainsfromcalves.JClin Microbiol.1998;36:1795–7.
31.WangG,ClarkCG,RodgersFG.DetectioninEscherichiacoliof thegenesencodingthemajorvirulencefactors,thegenes definingtheO157:H7serotype,andcomponentsofthetype2 ShigatoxinfamilybymultiplexPCR.JClinMicrobiol. 2002;40:3613–9.
32.MarzonyET,KamaliM,SaadatiM,KeihanAH,FooladiAAI, SajjadiS.SinglemultiplexPCRassaytoidentifytheshiga toxin.AfrJMicrobiolRes.2011;5:1794–800.
33.FachP,PerelleS,DilasserF,GroutJ.Comparisonbetweena PCR-ELISAtestandtheverocellassayfordetectingShiga toxin-producingEscherichiacoliindairyproductsand characterizationofvirulencetraitsoftheisolatedstrains.J ApplMicrobiol.2001;90:809–18.
34.GeB,ZhaoS,HallR,MengJ.APCR-ELISAfordetectingShiga toxin-producingEscherichiacoli.MicrobesInfect.