h ttp : / / w w w . b j m i c r o b i o l . c o m . b r /
Clinical
Microbiology
Identification
of
pathogenic
and
nonpathogenic
Leptospira
species
of
Brazilian
isolates
by
Matrix
Assisted
Laser
Desorption/Ionization
and
Time
Flight
mass
spectrometry
Daniel
Karcher
a,1,
Rafaella
C.
Grenfell
a,1,
Andrea
Micke
Moreno
c,
Luisa
Zanolli
Moreno
c,
Silvio
Arruda
Vasconcellos
c,
Marcos
B.
Heinemann
c,
Joao
N.
de
Almeida
Junior
b,
Luiz
Juliano
a,∗,
Maria
A.
Juliano
aaUniversidadeFederaldeSãoPaulo,EscolaPaulistaMedicina,DepartamentodeBiofísica,SãoPaulo,SP,Brazil
bUniversidadedeSãoPaulo,InstitutodeMedicinaTropicaldeSãoPauloLaboratóriodeMicologiaMédicaDivisãodeLaboratórioCentral
–LIM-03,HospitaldasClínicasdaFaculdadedeMedicinadaUniversidadedeSãoPaulo,SãoPaulo,SP,Brazil
cUniversidadedeSãoPaulo,FaculdadedeVeterináriaeZootecnia,DepartamentoMedicinaVeterináriaPreventivaeSaúdeAnimal,São
Paulo,SP,Brazil
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received20November2016 Accepted21March2018 Availableonline13April2018 AssociateEditor:RoxanePiazza
Keywords: Leptospira Brazil Identification Massspectrometry MALDI-TOF
a
b
s
t
r
a
c
t
MatrixAssistedLaserDesorption/IonizationandTimeofFlightmassspectrometry (MALDI-TOFMS)is apowerfultool forthe identificationofbacteria throughthedetectionand analysisoftheirproteinsorfragmentsderivedfromribosomes.Slightsequencevariationsin conservedribosomalproteinsdistinguishmicroorganismsatthesubspeciesandstrain lev-els.CharacterizationofLeptospiraspp.by16SRNAsequencingiscostlyandtime-consuming, andrecentstudieshaveshownthatcloselyrelatedspecies(e.g.,Leptospirainterrogansand Leptospirakirschneri)maynotbediscriminatedusingthistechnology.Herein,wereportan in-houseLeptospirareferencespectradatabaseusingLeptospirareferencestrainsthatwere validatedwithacollectionofwell-identifiedBrazilianisolateskeptintheBacterial Zoono-sisLaboratoryattheVeterinaryPreventiveMedicineandAnimalHealthDepartmentatSao PauloUniversity.Inaddition,L.interrogansandL.kirschneriweredifferentiatedusingan in-depthmassspectrometryanalysiswithClinProToolsTMsoftware.Inconclusion,our in-housereferencespectradatabasehasthenecessaryaccuracytodifferentiatepathogenic andnon-pathogenicspeciesandtodistinguishL.interrogansandL.kirschneri.
©2018SociedadeBrasileiradeMicrobiologia.PublishedbyElsevierEditoraLtda.Thisis anopenaccessarticleundertheCCBY-NC-NDlicense(http://creativecommons.org/ licenses/by-nc-nd/4.0/).
∗ Correspondingauthor.
E-mails:ljuliano@terra.com.br,juliano.epm@hotmail.com(L.Juliano). 1 Theseauthorscontributedequallytotheexecutionofthiswork.
https://doi.org/10.1016/j.bjm.2018.03.005
Introduction
LeptospirosisisamammalianzoonosiscausedbyLeptospira strains belonging to the order Spirochaetales. Mammals, including humans, are affected bydifferent clinical mani-festations,dependingonthe virulence,motility,and ability ofthe leptospiralpathogen to surviveinthe host. Suscep-tibility to infection is dependent on age, genetic factors and skin integrity during the infection. Leptospira biology andleptospirosisphysiopathologywerecomprehensively pre-sentedanddiscussedinarecentpublication.1Theantigenic diversityamongserovarsdifferentiatespathogenic(Leptospira interrogans) and non-pathogenic or saprophytic (Leptospira biflexa)species.2 Atleast22specieshavebeenclassified by moleculartechniques.2–4Themicroscopicagglutinationtest (MAT)isthemostcommonlyuseddiagnosticmethodinthe clinic; however, limitations have been previously reported anddiscussed.3,5ThecharacterizationofLeptospiraspp.using moleculartechniquessuchas16SRNAsequencingiscostly andtime-consuming,6especiallytakingintoaccountthelarge numberofmicroorganismsidentifiedintheclinicalpractice. Thismethoddependsononeorseveraltargetgenes,however thedataforallthepeptideswithamassrangeof2–20kDa couldbecollectedusingMALDI-TOFMSasdemonstratedby Xiaoetal.7onmolecularfingerprintingofpathogenicand non-pathogenic Leptospira. MALDI-TOF MS is a well-established techniquefortherapidcharacterizationofbacteria, andits useiscontinuouslyincreasing.8Thistechnologycan differen-tiatemicroorganisms’speciesbytheanalysisandcomparison ofproteinsorproteinfragmentsderivedfromribosomes.Itis importanttonotethatslightsequencevariationsinconserved ribosomalproteinsare sufficienttodistinguish microorgan-ismsatthesubspeciesandstrainlevels.8MALDI-TOFMShas been proposed tobe a powerful tool for the identification ofLeptospiraatthespecieslevel.6–8,10However,the misiden-tification ofL.interrogansas L.kirschneri byMALDI-TOFMS hasbeendescribed,and potentialbiomarkersto differenti-ate these species have been investigated.10 In the present paper,wefocusedon(i) thecharacterizationofpathogenic andnon-pathogenicLeptospiraspeciesofaLeptospira Brazil-iancollectionusingMALDI-TOFMSaftercreatinganin-house databaseand(ii)the differentiationofL.interrogansfrom L. kirschneribyin-depthmassspectrometryanalysis.
Material
and
methods
Leptospirastrainsandisolates
Thirty-onereferenceleptospiralstrainsand22fieldisolates belongingtopathogenic(Leptospirainterrogans,Leptospira borg-petersenii,Leptospirakirschneri,LeptospiranoguchiiandLeptospira santarosai)andnon-pathogenic(Leptospirabiflexa)specieswere analyzed.TheLeptospiraisolateswererecoveredfrombovine, dog,human,Rattusnorvegicus,andRattusrattusurinesamples takenfromSaoPaulo,RiodeJaneiroandLondrina(Table1). ThestrainsandisolatesweremaintainedintheLaboratoryof BacterialZoonosis–SchoolofVeterinaryMedicineandAnimal Science/UniversityofSaoPaulo(USP)andstoredinFletcher
semi-solidmedium(FletcherMediumBase,DifcoTM,NJ,USA) at30◦C.Thespeciesofthefieldisolateswerepreviously
iden-tifiedby16SrRNAsequencing(datanotshown).
SamplepreparationforMALDI-TOFanalysis
The strains and isolateswere grownand diluted (1:25)for seven days at 30◦C in
Ellinghausen-McCullough-Johnson-Harris medium(EMJHDifcoTM,NJ, USA),andbacterial cells werecountedusingaPetroffHaussercountingchamber(HS Hausser Scientific, Horsham, PA) bydark field microscopy. Leptospira cultureswerecentrifuged at11,000×gfor10min atroomtemperature,andthepelletwaswashedtwicewith 3mLof phosphate buffered saline (PBS) and suspendedin steriledeionized watertoa finalbacterialconcentrationof 1×108organismspermL.Ethanol/formicacidprotein extrac-tionwasperformedbyadditionof300Lofthecultureinto 900Lofethanol(99.8%,PA)followedvortexingand10-min ofincubation. This inactivationprocedure was followedby a 10-mincentrifugation at11,000×g atroom temperature, the supernatantwasremoved,andthepelletwas airdried untiltheethanolwascompletelyevaporated.Thisprocesswas repeatedandthematerialwasthendissolvedin30Lof70% formicacid(Sigma–Aldrich)followedbytheadditionof30L ofacetonitrile(FlukaAnalyticalSigma–Aldrich,Munich, Ger-many).Centrifugationwasperformedat11,000×gfor2min atroomtemperature.Twomicrolitersoftheclearsupernatant werespottedona384targetpolishedsteelplate(Bruker Dal-tonikGmbH,Bremen,Germany)andallowedtodry.Following this, the driedspot was overlaid with2Lofmatrix solu-tion,asaturatedsolutionof␣-Cyano-4-hydroxycinnamicacid (HCCA,99%BrukerDaltonikGmbH,BremenorSigma–Aldrich, Munich,Germany)(10mg/mL)inacetonitrile(FlukaAnalytical Sigma–Aldrich)and0.1%trifluoraceticacid(1:2)(TFA-Reagent PlusW99%,Sigma–Aldrich).Finally,sampleswereallowedto dry atroomtemperature.Escherichia coliDH5␣ wasusedas apositivecontrol,andanon-inoculatedmatrixsolutionwas used asa negativecontrol.During data acquisition,it was observedthatsomeisolatesunderwentosmoticlysisin deion-izedwater,whichwascorrectedbyreplacingsteriledeionized waterbysalinesolution(0.85%NaCl)bufferedwithSorensen’s solution(69mMNa2HPO4/8mMNaH2PO4,pH7.6).9This solu-tion has lower osmolarity than PBS, but kept cells intact withoutinterferingwiththeionizationofthebacterial pro-teinsaswellasthemassfingerprintofourpreviouslydata thatweregeneratedinsalinesolution.Additionalmass spec-trawerethenobtainedwithfreshculturepassagestoensure theminimumnumberofspectraforthegenerationofsingle MainSpectrumProfiles(MSP).
InstrumentsettingsforMALDI-TOFMSanalysis
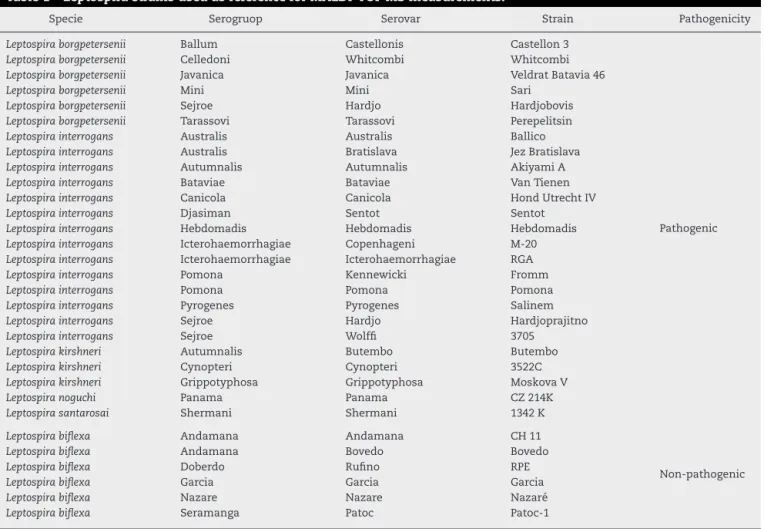
Table1–LeptospirastrainsusedasreferenceforMALDI-TOFMSmeasurements.
Specie Serogruop Serovar Strain Pathogenicity
Leptospiraborgpetersenii Ballum Castellonis Castellon3
Pathogenic
Leptospiraborgpetersenii Celledoni Whitcombi Whitcombi
Leptospiraborgpetersenii Javanica Javanica VeldratBatavia46
Leptospiraborgpetersenii Mini Mini Sari
Leptospiraborgpetersenii Sejroe Hardjo Hardjobovis
Leptospiraborgpetersenii Tarassovi Tarassovi Perepelitsin
Leptospirainterrogans Australis Australis Ballico
Leptospirainterrogans Australis Bratislava JezBratislava
Leptospirainterrogans Autumnalis Autumnalis AkiyamiA
Leptospirainterrogans Bataviae Bataviae VanTienen
Leptospirainterrogans Canicola Canicola HondUtrechtIV
Leptospirainterrogans Djasiman Sentot Sentot
Leptospirainterrogans Hebdomadis Hebdomadis Hebdomadis
Leptospirainterrogans Icterohaemorrhagiae Copenhageni M-20
Leptospirainterrogans Icterohaemorrhagiae Icterohaemorrhagiae RGA
Leptospirainterrogans Pomona Kennewicki Fromm
Leptospirainterrogans Pomona Pomona Pomona
Leptospirainterrogans Pyrogenes Pyrogenes Salinem
Leptospirainterrogans Sejroe Hardjo Hardjoprajitno
Leptospirainterrogans Sejroe Wolffi 3705
Leptospirakirshneri Autumnalis Butembo Butembo
Leptospirakirshneri Cynopteri Cynopteri 3522C
Leptospirakirshneri Grippotyphosa Grippotyphosa MoskovaV
Leptospiranoguchi Panama Panama CZ214K
Leptospirasantarosai Shermani Shermani 1342K
Leptospirabiflexa Andamana Andamana CH11
Non-pathogenic
Leptospirabiflexa Andamana Bovedo Bovedo
Leptospirabiflexa Doberdo Rufino RPE
Leptospirabiflexa Garcia Garcia Garcia
Leptospirabiflexa Nazare Nazare Nazaré
Leptospirabiflexa Seramanga Patoc Patoc-1
CollectionattheBacterialZoonosesLaboratory,DepartmentofVeterinaryPreventiveandAnimalHealthofSchoolofVeterinaryMedicineand AnimalScience,SãoPauloUniversity,Brazil.
Allspectrawere analyzedbystandardpattern-matching algorithmusingtheMALDIBiotyperTM 3.1software(Bruker
Daltonics), and the raw spectra were compared with the reference spectra of the Bruker library (database version 3.3.1, 5627reference spectra)with defaultsettings. TheID criteria used was the recommended by the manufacturer: – a score ≥2.000 indicated species, a score between 1.700 and 1.999 indicated genus level and a score <1.700 was interpreted as no ID. For MainSpectra (MSP) and dendro-gramconstruction,flat-liners and bad qualityspectrawere removedand additionalmeasurementswere carriedout to obtain20spectrafromeachisolate/strain.Spectrawerethen loaded into BiotyperTM 3.1 software (Bruker Daltonics) for
MSPcreationand dendrogramclustering construction with thedefaultsettings(distance measure:correlation;linkage: average; scoreoriented). Each spot was measured in 1000-shotstepsforatotalof4000lasershots.Preparationofthe BTSandcalibrationwereperformedfollowingthe manufac-turer’sinstructions:calibrationwassuccessfulwhenproteins of the mass spectra were in a range of ±200 parts per million(ppm).
In-housedatabaseanddendrogramconstruction
Foreachofthe31strains,30individualspectrawereusedto createaMSP.Flat-linersandbadqualityspectrawereremoved, and additional measurements were carried out to obtain 30 spectrafrom each isolate/strain.TheMSP wasobtained usingMALDI-Biotypersoftware(BrukerDaltonics,Germany) andthenloadedintotheBrukerDaltonicsdatabase(version 3.1.2.0).SoftwaresettingsforMSPcreationweresetto max-imalmasserrorofeachsinglespectrum:2000;desiredmass errorfortheMSP:200;desiredpeakfrequencyminimum(%): 25;andmaximaldesiredpeaknumberoftheMSP:70. Den-drogramclusteringwasconstructedwiththedefaultsetting of160(distancemeasure:correlation;linkage:average;score oriented).
Determiningtheefficiencyofthedatabasesearchwith Leptospirafieldisolates
wereexpressedinlogscorevalues,withvalues≥2 indicat-ingreliablespeciesidentificationandvaluesfrom1.7to2.00 indicatingreliablegenusidentification.
DifferentiationofLeptospirainterrogansandLeptospira
kirschneriusingClinProToolsTM
ClinProToolsTM (BrukerDaltonics)generatesmultiple math-ematicalalgorithmstogeneratepatternrecognitionmodels for the classification and prediction of different classes (e.g., L. interrogans class 1, L. kirschneri class 2) from mass spectrometry-basedprofilingdata.Variousspectraofthe dif-ferentserovars(03serovartoL.kirschneriand12 serovarto L.interrogans)wereusedforeachclass,seekingto standard-izethedataforspeciesdistinction.Moreover,ClinProToolsTM provides a list of peaks sorted according to the statistical significance to differentiate between both classes.12 Thus, torecognizemassspectrapatternsandbiomarkersbetween L.interrogansandL.kirschneri,spectrapeakanalysismodels with ClinProToolsTM software v.3.0 (Bruker Daltonics) were created from anadditional 210mass spectraof11 L. inter-rogans (15 high-quality mass spectra per isolate) and 3 L. kirschneri(15high-qualitymassspectraperisolate)isolates. Spectrawerepretreatedwitharesolutionof800ppm,amass rangeof2000–20,000Da,atophatbaselinesubtractionwith 10%minimalbaselinewidth,enablingnullspectraexclusion, recalibrationwith500ppmmaximalpeakshiftand30%match celebrant peaks. ClinProToolsTM models (Bruker Daltonics) were generated using three algorithms: Genetic Algorithm (GA),SupervisedNeuralNetwork(SNN),andQuickClassifier (QC).Foreachmodel,therecognitioncapability(RC)andcross validation(CV)percentageweregeneratedtodemonstratethe reliabilityandaccuracyofthemodel.RCandCVpercentages areindicatorsofthemodel’sperformanceandserveasuseful predictorsofthemodel’sabilitytoclassifytestisolates.We alsocarriedoutprincipalcomponentanalysis(PCA)included inClinProToolssoftwareaimingtovisualizehomogeneityand heterogeneityofthe proteinspectraofL.interrogansand L. kirschneri.Principalcomponentanalysis(PCA)andtheresults wereshownin3Dscoreplot.
Single-peakanalysis
Foreachpeak,theAUCforthediscriminationofthegroups wasdirectlyobtainedfromtheClinProToolsTMv.3.0software (BrukerDaltonics).ForthefivepeakswiththehighestAUC,the detection performanceswere verified usingFlexAnalysisTM v.3.4(BrukerDaltonics). Aftersmoothing and baseline sub-traction,themasslistsforeachisolatewereobtainedusing thecentroidalgorithmwithasignal-to-noise(SN)threshold of0.5andamaximumof500peaksandexportedtoMicrosoft Excel.TheSNratiosofthepeakswithatoleranceof1000ppm were exported toSPSS version 18.0.ROC (Receiver Operat-ingCharacteristic)curveswereconstructed,andtheiroptimal cutoffvalueswere determined withthemaximum Youden index.11
Results
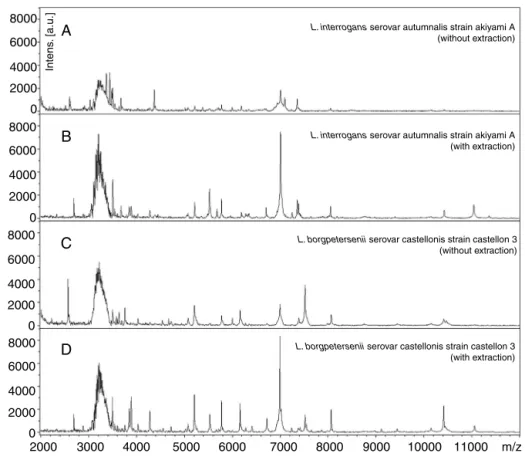
Referencespectrawerecreated forall31 leptospiralstrains andappliedasunassignedMSPsinthecommercially avail-ableMALDIBiotyperTMdatabasespectralibrary,whichlacks leptospiralproteinprofiles(Fig.1).TheMSPswereclustered accordingtopathogenicityintheMALDI-TOFMSdendrogram, and the pathogenic species (red) are clearly differentiated fromthenonpathogenicLeptospiraspecies(green)(Fig.2). Sim-ilarly, the pathogenic L. borgpeterseniiand L. interrogansare locatedinseparateclusters,but,asexpected,poor discrim-inationwasobtainedforL.interrogansandL.kirschneri.
RepresentativemassspectraofL.interrogansand L. borg-peterseniiobtainedbydirectanalysisandbyproteinextraction protocolareshowninFig.1.InAandC,massspectrawere obtainedwithoutproteinextractionandpeakswithlow inten-sitywereobserved.Incontrast,BandDshowhigherquality massspectraobtainedafterproteinextractionprotocol,with peakswithhigherintensity.
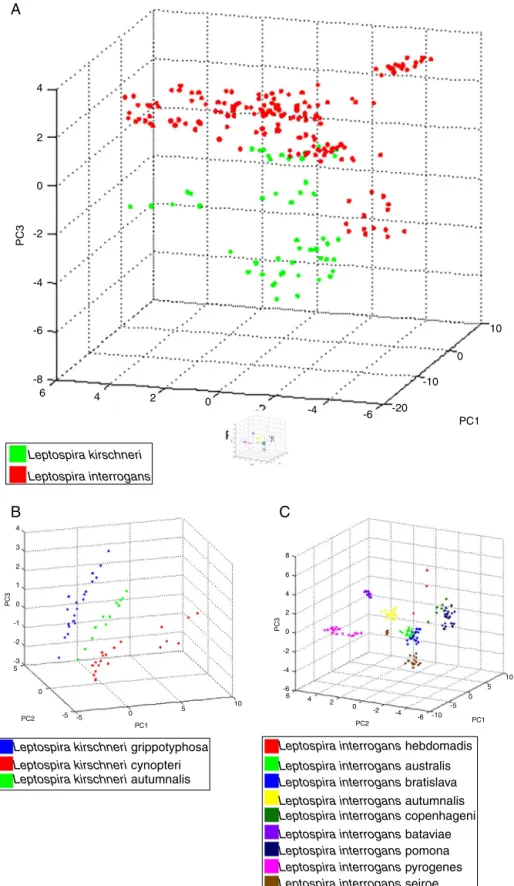
The 22 field isolates belonging to L. biflexa, L. interro-gans and L. santarosai had the correct species assigned by MALDI-TOFMS,andallisolatesshowedscorevaluesover2.0 (Table2),whereit ispossibletoidentifyall isolatesbythe correctspeciesIDfollowingourin-housedatabase. ThePCA reproduces through different statistical teststhe reduction ofseveral variablesofaset ofdata,whereeach point rep-resentsaspectrumandeachcolorrepresentsagroupingof similardata.Fig.3ApresentsthePCAforL.interrogansspecies inredandL.kirschneriingreen,thereisaperceptible distinc-tion betweenthetwogroupsevenwithsomecloserpoints showing that the PCAanalysis does not guarantee aclear separationbetweenthespecies.BpresentsthePCAforthe serovarsthatformedtheclassL.kirschneriinA,aclear sepa-rationbetweentheserovarsisobserved.CpresentsthePCA fortheserovarsthatformtheclassL.interrogans,whichshows thatthegrouprepresentingL.interrogansserovarBataviaecan becompletelyseparated,sincetheotherclusterscannotbe separated.
The three classification models from ClinProToolsTM showed RCvalues ≥90%inthe discrimination ofL. interro-gansandL.kirschneri.Thebestresultswereprovidedbythe GAmodel,withRCandCVvaluesof100%and97%, respec-tively.DetailsofthesevaluesareshowninTable3.Thestrain distributionmapsbasedontheGAmodelshowthatL. inter-rogans andL.kirschnericanbedistinguishedbased ontheir peptide massfingerprints,thebest separatingpeaksofthe currentstatisticsortorderaredisplayedinFig.4.
A
B
C
D
8000 8000
L. borgpetersenii serovar castellonis strain castellon 3 (with extraction) L. borgpetersenii serovar castellonis strain castellon 3 (without extraction) L. interrogans serovar autumnalis strain akiyami A (with extraction) L. interrogans serovar autumnalis strain akiyami A (without extraction)
8000
8000
8000 0
0
0
0 2000
2000
2000
2000
2000 3000
4000
4000
4000
4000
4000 5000 7000 9000 10000 11000 m/z
6000
6000
6000
6000
6000
Intens
. [a.u.]
Fig.1–MALDI-TOFMSspectraobtainedbyanalyzingthereferencestrainsofLeptospirainterrogansandLeptospira
borgpeterseniiwithandwithoutextractionasdescribedin“Materialandmethods”section.Thesedatashowtheimportance oftheproteinextractiontoobtainthebetterqualityofspectra.
MSP dendogram
Leptospira biflexa sg. saramanga sv. patoc strain patoc 1 Leptospira biflexa sg. andamana sv. bovedo strain bovedo
Leptospira kirchneri sg. grippotyphosa sv. grippotyphosa strain moskva V Leptospira interrogans sg. autumnalis sv. autumnalis c. akiyami A
Leptospira kirschneri sg. cynopteri sv. cynopteri strain 3522 C Leptospira kirschneri sg. autumnalis sv. butembo strain butembo Leptospira interrogans sg. australis sv. bratislava strain jez bratislava Leptospira interrogans sg. australis sv. australis c. ballico
Leptospira interrogans sg. canicola sv. canicola strain hond uthecht IV Leptospira interrogans sg. pomona sv. pomona strain pomona Leptospira interrogans sg. hebdomadis sv. hebdomadis strain hebdomadis Leptospira interrogans sg. icterohaemorrhagiae sv. copenhageni strain M 20 Leptospira interrogans sg. sejroe sv. hardjo c. hardjoprojitno
Leptospira interrogans sg. sejroe sv. wolfii c. 3705 Leptospira noguchii sg. pamana sv. pamana strain CZ 214K Leptospira borgpetersenii sg. celledoni sv. whitcombi strain whitcombi Leptospira borgpetersenii dg. mini sv.mini strain sari
Leptospira borgpetersenii sg. javanica sv. javanica strain veldrat batavia 46 Leptospira santarosai sg. shemani sv. shemani c. 1342 k
Leptospira biflexa sg. andamana sv. andamanda strain ch-11
Distance level
0 100 200 300 400 500 600
0 0
2
2
0 10
PC1 -10
-20 -2
4
-8 -6
-6 6
-4
-4 4
A
C
5
0 0
PC3
PC3
PC3
PC2 PC2
PC1 PC1
10
10
-10 5
5
-5 -5
-5 -3 -2 -1 0 1 2 3 4
-2
-2 -6
-6 6
-4
-4 4
0
0 0
2
2 4 6 8
B
Leptospira interrogans
Leptospira interrogans sejroe
Leptospira interrogans pyrogenes
Leptospira interrogans pomona
Leptospira interrogans bataviae
Leptospira interrogans copenhageni
Leptospira interrogans autumnalis
Leptospira interrogans bratislava
Leptospira interrogans australis
Leptospira interrogans hebdomadis
Leptospira kirschneri
Leptospira kirschneri autumnalis
Leptospira kirschneri cynopteri
Leptospira kirschneri grippotyphosa
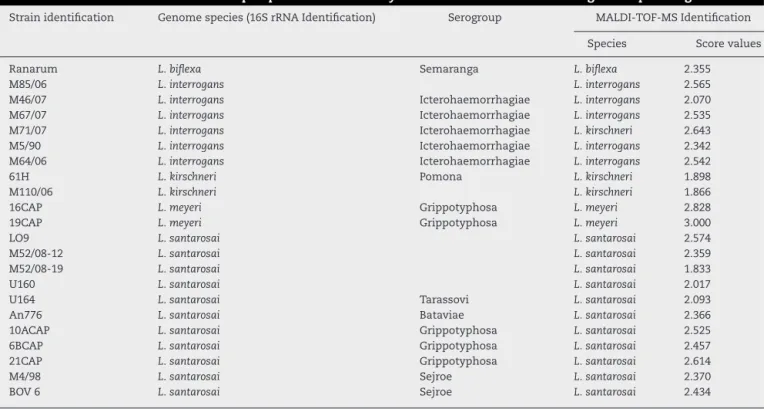
Table2–Identificationresultsof22leptospiralfieldisolatesbyMALDI-TOFMSand16SrRNAgenesequencing.
Strainidentification Genomespecies(16SrRNAIdentification) Serogroup MALDI-TOF-MSIdentification
Species Scorevalues
Ranarum L.biflexa Semaranga L.biflexa 2.355
M85/06 L.interrogans L.interrogans 2.565
M46/07 L.interrogans Icterohaemorrhagiae L.interrogans 2.070
M67/07 L.interrogans Icterohaemorrhagiae L.interrogans 2.535
M71/07 L.interrogans Icterohaemorrhagiae L.kirschneri 2.643
M5/90 L.interrogans Icterohaemorrhagiae L.interrogans 2.342
M64/06 L.interrogans Icterohaemorrhagiae L.interrogans 2.542
61H L.kirschneri Pomona L.kirschneri 1.898
M110/06 L.kirschneri L.kirschneri 1.866
16CAP L.meyeri Grippotyphosa L.meyeri 2.828
19CAP L.meyeri Grippotyphosa L.meyeri 3.000
LO9 L.santarosai L.santarosai 2.574
M52/08-12 L.santarosai L.santarosai 2.359
M52/08-19 L.santarosai L.santarosai 1.833
U160 L.santarosai L.santarosai 2.017
U164 L.santarosai Tarassovi L.santarosai 2.093
An776 L.santarosai Bataviae L.santarosai 2.366
10ACAP L.santarosai Grippotyphosa L.santarosai 2.525
6BCAP L.santarosai Grippotyphosa L.santarosai 2.457
21CAP L.santarosai Grippotyphosa L.santarosai 2.614
M4/98 L.santarosai Sejroe L.santarosai 2.370
BOV6 L.santarosai Sejroe L.santarosai 2.434
CollectionattheBacterialZoonosesLaboratory,DepartmentofVeterinaryPreventiveandAnimalHealthofSchoolofVeterinaryMedicineand AnimalScience,SãoPauloUniversity,Brazil.
Table3–Completeresultsderivedfromtheclassificationmodels.
Classificationmodel Crossvalidation(CV)(%)Recognitioncapability(RC)(%) Integrationregionsusedforclassification
Peak#1(Da)Peak#2(Da)Peak#3(Da)Peak#4(Da)Peak#5(Da)
GAa 97.2 100.0 8057 4671 5472 8084 8305
SNNb 55.6 100.0 8057 8097 6710 8084 12,679
QCc 92.6 93.7 8057 – – – –
ResultsobtainedbyanalyzingofClinProTools. a GeneticAlgorithm.
b SupervisedNeuralNetwork. c QuickClassifier.
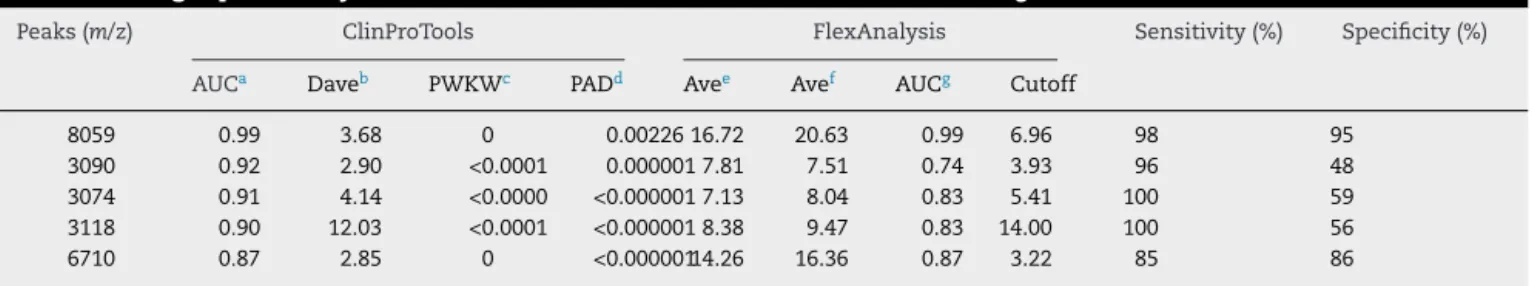
andsingle-peakanalysisresultsforthe differentiationofL. interrogansfromL.kirschneriare summarizedinTable4and exemplifiedinFig.5.
Discussion
Duringleptospirosisoutbreaks,Leptospiraspecies identifica-tion is an essential step for tracking and controlling the pathogentransmission.Thedeterminationofaserovarmay be insufficient as different species may have the same serovarbut may be distinct intheirability tocause mam-malianinfection.13DNAsequencingisanalternativemethod
Table4–Single-peakanalysisforthediscriminationofL.kirschneriandL.interrogans.
Peaks(m/z) ClinProTools FlexAnalysis Sensitivity(%) Specificity(%)
AUCa Daveb PWKWc PADd Avee Avef AUCg Cutoff
8059 0.99 3.68 0 0.00226 16.72 20.63 0.99 6.96 98 95
3090 0.92 2.90 <0.0001 0.0000017.81 7.51 0.74 3.93 96 48
3074 0.91 4.14 <0.0000 <0.0000017.13 8.04 0.83 5.41 100 59
3118 0.90 12.03 <0.0001 <0.0000018.38 9.47 0.83 14.00 100 56
6710 0.87 2.85 0 <0.00000114.26 16.36 0.87 3.22 85 86
PeakswiththebestperformancesaccordingtoClinProToolsTMandFlexAnalysisTMsoftwares. a AUC,areaunderthecurve.
b Dave,differencebetweenthemaximalandtheminimalaveragepeakarea/intensityofthegroups. c PWKW,p-valueofWilcoxon/Kruskal–Wallistest(range:0–1;0Dgood).
d PAD:p-valueofAnderson–Darlingtest,<0.05indicatesdatanotnormallydistributed;givesinformationaboutthenormaldistribution(range: 0–1;0=notnormallydistributed).
e Ave,area/intensityaverageofagroupfromLeptospirakirschneri. f Ave,area/intensityaverageofagroupfromLeptospirainterrogans.
g AUCsandsignal-to-noisecutoffvalueswereobtainedfromanROCcurveconstructedusingSPSSVersion18.0andFlexAnalysis.
byothercentersthatalsoconstructedinhouseLeptospiraMSP databases.Moreover,allfieldisolateshadthecorrectspecies assigned,withscores above2.0,which ensuresthe quality ofour MSP database forLeptospira species ID.The distinc-tionofL.interrogansandL.kirschneriusingClinProToolsTMand single-peakanalysisisalsonoteworthy.AlthoughMALDI-TOF MShasalreadybeensuccessfullyappliedinLeptospiragenus andspeciesidentification,6–8 themisidentificationofclosely relatedspecies,suchasL.interrogansandL.kirschneri,hasalso beenreportedandrepresentsanimportantchallengeinthe implementationofMALDI-TOFMSforLeptospiraidentification. Hereweobservedthatwithproperanalysis,Leptospiraspecies canbedistinguishedbasedontheirpeptidemassfingerprints. The ClinProToolsTM software is a biomarker analyzer that has been widely applied in microbiology, providing a rapidandcost-savingmethodforepidemiologicalclustering, strain typing and subspeciesidentification.14–16 Using both ClinProToolsTMandsingle-peakanalysiswithFlexAnalysisTM has provided higher discriminatory power to detect bio-markerpeaks.14–17 Ourresultscorroboratepreviousfindings thatoneisolatebiomarkerwith8000–8100Dacaneffectively distinguishthe closelyrelatedpathogenicspeciesL. interro-gansfromL.kirschneri.10Indeed,wefurtherdescribedtheSN cut-offvaluethathastobeadoptedtoaccuratelydifferentiate thesetwotaxabyasimpleinspectionofthemassspectrum. Recently,L.kirschneriserovarMosdokwas,forthefirsttime, linkedtohumanleptospirosisinthe southernhemisphere; therefore,rapidspeciesIDusingMALDI-TOFMSmaybethe firststeptoimplementcontrolstrategies.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest,evenduringthe itemproofs.
Pk 6,711 Da
Pk 8,057 Da
8.0 9.0
0.0 0.0 1.0
1.0 2.0
2.0 3.0
3.0 4.0
4.0 5.0
5.0 6.0
7.0
6.0 7.0 0.5
0.5 1.5
1.5 2.5
2.5 3.5
3.5 4.5
4.5 5.5
5.5 6.5
6.5 7.5
8.5
Fig.4–StraindistributionmapcorrespondingtoLeptospira interrogans(red)andLeptospirakirschneri(green).Thex-axis showsthepeakarea/intensityvalueswithrespecttothe mostrelevantpeak(8057Da)todistinguishLeptospira interrogans(red)fromLeptospirakirschneri(green).The
y-axisshowsthepeakarea/intensityvalueswithrespectto thepeakwith(6711Da)fromLeptospirainterrogans(red)and
Leptospirakirschneri(green).Theellipsesrepresentthe spectrawithgreaterdistinctionbetweenthetwogroups, whereasprominentpeaksinthex-axisandy-axis.
Acknowledgements
0
0 1000
1000 2000
7950 8000 8150 m/z
Leptospira interrogans
Leptospira kirschneri
8250 8200
8100 8050
200 400 600 800 3000 4000
8098
Intens
. [a.u.]
Intens
. [a.u.]
8059
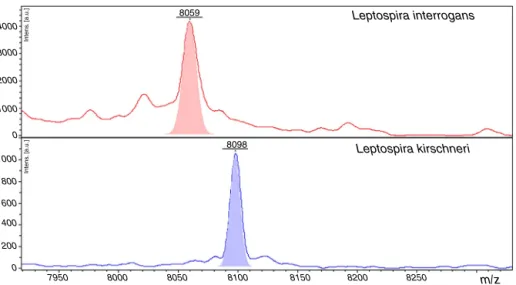
Fig.5–RepresentativespectraofLeptospirainterrogansandLeptospirakirschineri.Therepresentativepeaksthatallow differentiationofthestrainsinthespectraareshowed,inredforL.interrogans,andinblueforL.kirschneri.Thepeakwith
m/z=8059inL.interroganswedetectedasshowninTable3byClinProToolsanalysis.Thepeakm/z=8098inL.kirschineri
waspreviouslyidentifiedbyRettingeretal.10
eTecnologico(CNPq—Projects– 443978-2014-0and 467478-2014-7).L.Z.M.isrecipientofaPhDfellowshipfromFAPESP (2013/17136-2).
r
e
f
e
r
e
n
c
e
s
1. HaakeDA,LevettPN.Leptospirosisinhumans.CurrTop MicrobiolImmunol.2015;387:65–97.
2. EvangelistaKV,CoburnJ.Leptospiraasanemerging pathogen:areviewofitsbiology,pathogenesisandhost immuneresponses.FutureMicrobiol.2010;5(9):1413–1425.
3. CerqueiraGM,PicardeauM.AcenturyofLeptospirastrain typing.InfectGenetEvolJMolEpidemiolEvolGenetInfectDis. 2009;9(5):760–768.
4. LilenbaumW,KremerF,RistowP,etal.Molecular
characterizationofthefirstleptospiresisolatedfromgoats inBrazil.BrazJMicrobiolPublBrazSocMicrobiol.
2014;45(4):1527–1530.
5. MillerMD,AnnisKM,LappinMR,LunnKF.Variabilityin resultsofthemicroscopicagglutinationtestindogswith clinicalleptospirosisanddogsvaccinatedagainst leptospirosis.JVetInternMedAmCollVetInternMed. 2011;25(3):426–432.
6. CalderaroA,PiccoloG,GorriniC,etal.Leptospiraspecies andserovarsidentifiedbyMALDI-TOFmassspectrometry afterdatabaseimplementation.BMCResNotes.2014;7:330.
7. DjelouadjiZ,RouxV,RaoultD,KodjoA,DrancourtM.Rapid MALDI-TOFmassspectrometryidentificationofLeptospira
organisms.VetMicrobiol.2012;158(1–2):142–146.
8. RettingerA,KrupkaI,GrünwaldK,etal.Leptospiraspp. strainidentificationbyMALDITOFMSisanequivalenttool to16SrRNAgenesequencingandmultilocussequence typing(MLST).BMCMicrobiol.2012;12:185.
9. DibCC,Gonc¸alesAP,deMoraisZM,etal.Cross-protection betweenexperimentalanti-leptospirosisbacterins.BrazJ MicrobiolPublBrazSocMicrobiol.2014;45(3):1083–1088.
10.KetterlinusR,HsiehS-Y,TengS-H,LeeH,PuschW.Fishing forbiomarkers:analyzingmassspectrometrydatawiththe newClinProToolssoftware.BioTechniques.2005;(Suppl.):37–40.
11.RuoppMD,PerkinsNJ,WhitcombBW,SchistermanEF. Youdenindexandoptimalcut-pointestimatedfrom observationsaffectedbyalowerlimitofdetection.BiomJ BiomZ.2008;50(3):419–430.
12.BrennerDJ,KaufmannAF,SulzerKR,SteigerwaltAG,Rogers FC,WeyantRS.FurtherdeterminationofDNArelatedness betweenserogroupsandserovarsinthefamily
LeptospiraceaewithaproposalforLeptospiraalexanderisp. nov.andfournewLeptospiragenomospecies.IntJSyst Bacteriol.1999;49Pt2:839–858.
13.XiaoD,ZhaoF,ZhangH,MengF,ZhangJ.Novelstrategyfor typingMycoplasmapneumoniaeisolatesbyuseof
matrix-assistedlaserdesorptionionization-timeofflight massspectrometrycoupledwithClinProTools.JClin Microbiol.2014;52(8):3038–3043.
14.ZhangT,DingJ,RaoX,etal.Analysisofmethicillin-resistant
Staphylococcusaureusmajorclonallineagesby
Matrix-AssistedLaserDesorptionIonization-TimeofFlight MassSpectrometry(MALDI-TOFMS).JMicrobiolMethods. 2015;117:122–127.
15.GrenfellRC,daSilvaJuniorAR,DelNegroGMB,etal. IdentificationofCandidahaemuloniicomplexspecies:useof ClinProTools(TM)toovercomelimitationsoftheBruker Biotyper(TM),VITEKMS(TM)IVD,andVITEKMS(TM)RUO Databases.FrontMicrobiol.2016;7:940.
16.AngelettiS,DicuonzoG,LoPrestiA,etal.MALDI-TOFmass spectrometryandblakpcgenephylogeneticanalysisofan outbreakofcarbapenem-resistantK.pneumoniaestrains.New Microbiol.2015;38(4):541–550.
17.daCunhaCEP,FelixSR,NetoACPS,etal.Infectionwith