An integration of genome-wide association study and gene expression profiling to prioritize the discovery of novel susceptibility Loci for osteoporosis-related traits.
Texto
Imagem
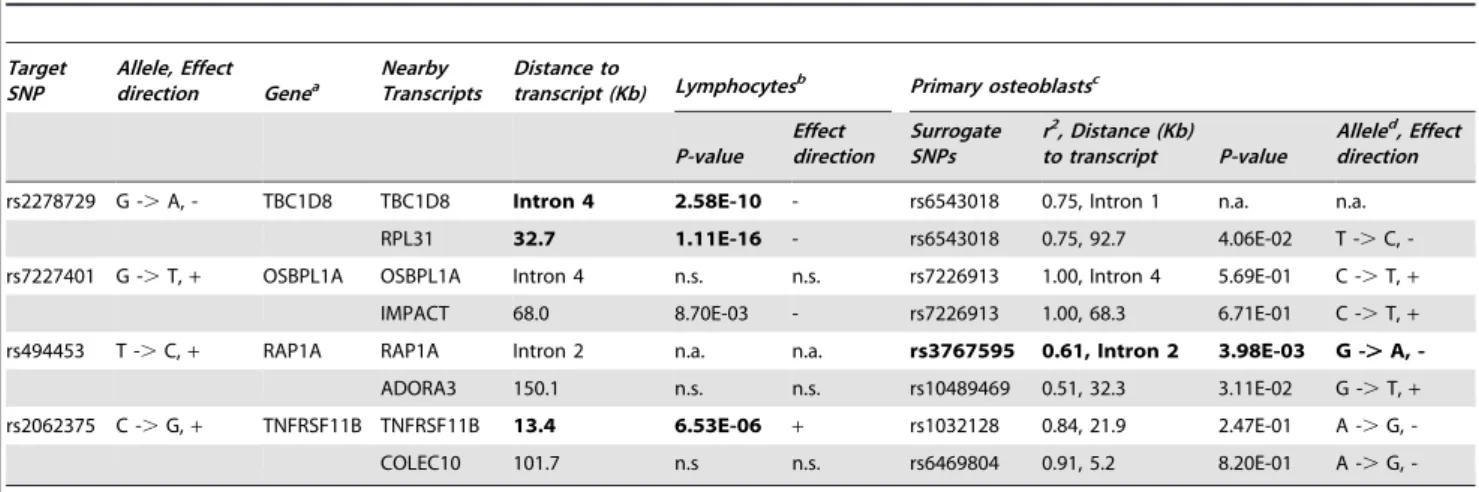
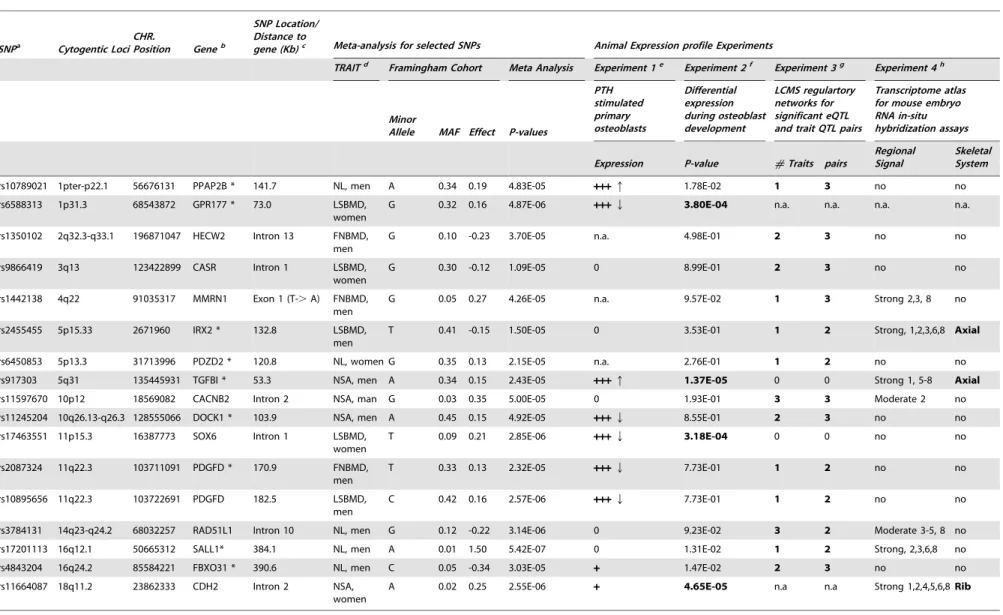
Documentos relacionados
The integration of functional gene annotations in the analysis confirms the detected differences in gene expression across tissues and confirms the expression of porcine genes being
***According to the following keys: AI: Aluminum induces expression of the gene; AR: Aluminum repress expression of the gene; DIS: deletion of the gen e increased susceptibility
We illustrate the integration problem and the generic flow approach with a real example found in studies of mouse gene expression within a large cooperative project between
Genome wide expression data from human psoriatic skin samples ( n = 58 patients) and normal skin ( n = 64 subjects) was analyzed to identify (A) the 50 genes most strongly increased
In summary, our data show that STC-1 mRNA and protein were expressed in the bovine osteoblasts, suggesting that STC-1 may affect bovine osteoblast differentiation and formation
To perform association tests between gene expression variation and SNP variation, we selected 374 of the 630 tested genes that had probe hybridization signals significantly above
For the unsupervised analysis and the supervised analyses (except that centered on the mitotic index), samples were divided in a learning set including our series pooled with West
Lists of genes analyzed using DAVID included genes over- expressed or under-expressed in all neutrophil populations compared to all non-neutrophil populations; genes up- or