www.bjorl.org
Brazilian
Journal
of
OTORHINOLARYNGOLOGY
ORIGINAL
ARTICLE
Prevalence
of
mitochondrial
DNA
mutations
in
sporadic
patients
with
nonsyndromic
sensorineural
hearing
loss
夽
Hua
Jiang,
Jia
Chen,
Ying
Li,
Peng-Fang
Lin,
Jian-Guo
He,
Bei-Bei
Yang
∗DepartmentofOtolaryngology,2ndAffiliatedHospital,SchoolofMedicine,ZhejiangUniversity,Zhejiang,China
Received29March2015;accepted6June2015 Availableonline5November2015
KEYWORDS
Mitochondria; rRNA; tRNA; Hearingloss
Abstract
Introduction:SeveralmitochondrialDNAmutationshavebeenreportedtobeassociatedwith nonsyndromichearinglossinseveralfamilies.However,littleisknownabouttheprevalenceof thesemutationsinsporadicpatientswithnonsyndromicsensorineuralhearingloss.
Objective: Thepurposeofourstudywastoinvestigatetheincidenceofthesemitochondrial DNAmutationsinsuchpopulation.
Methods:Atotalof178sporadicpatientswithnonsyndromicsensorineuralhearinglosswere enrolled in this study.Genomic DNA was extracted from the peripheral bloodsample. We employed the SNaPshot® sequencing method to detect five mitochondrial DNA mutations,
includingA1555GandA827Gin12SrRNAgeneandA7445G,7472insC,andT7511CintRNASer(UCN)
gene.Meanwhile,weusedpolymerasechainreactionandsequencedtheproducts toscreen GJB2genemutationsinpatientscarryingmitochondrialDNAmutations.
Results:WefailedtodetectthepresenceofA1555Gmutationin12SrRNAgene,andofA7445G, 7472insC,T7511CmutationsintRNASer(UCN)geneinourpopulation.However,wefoundthat6
patients(3.37%)werecarriersofahomozygousA827Gmutationandoneofthemalsocarried homozygousGJB2235delCmutation.
Conclusion: Ourfindingsinthepresentstudyindicatethateveninsporadicpatientswith non-syndromicsensorineuralhearingloss,mitochondrialDNAmutationsmightalsocontributetothe clinicalphenotype.
© 2015 Associac¸˜ao Brasileira de Otorrinolaringologia e Cirurgia C´ervico-Facial. Published by Elsevier Editora Ltda. This is an open access article under the CC BY license (http:// creativecommons.org/licenses/by/4.0/).
夽 Pleasecitethisarticleas:JiangH,ChenJ,LiY,LinP-F,HeJ-G,YangB-B.PrevalenceofmitochondrialDNAmutationsinsporadicpatients
withnonsyndromicsensorineuralhearingloss.BrazJOtorhinolaryngol.2016;82:391---6.
∗Correspondingauthor.
E-mail:yangbb1959@sina.com(B.-B.Yang).
http://dx.doi.org/10.1016/j.bjorl.2015.06.006
PALAVRAS-CHAVE
Mitocôndrias; rRNA; tRNA;
Deficiênciaauditiva
Prevalênciademutac¸õesnoDNAmitocondrialempacientesesporádicoscom deficiênciaauditivasensorioneuralnãosindrômica
Resumo
Introduc¸ão:Diversasmutac¸õesdoDNAmitocondrialtemsidodescritas,emdiferentesfamílias, associadasàdeficiênciaauditivanãosindrômica.Noentanto,poucosesabesobrea prevalên-ciadessas mutac¸ões em pacientesesporádicos comdeficiência auditivasensorioneural não sindrômica.
Objetivo:A finalidade donosso estudo foi investigar aincidência dessas mutac¸ões no DNA mitocondrialnessapopulac¸ão.
Método: Nototal,178pacientesesporádicoscomdeficiênciaauditivasensorioneuralnão sin-drômicaforamrecrutadosparaparticipac¸ãonoestudo.ODNAgenômicofoiextraídodeamostra desangueperiférico.Utilizamos ométodode sequenciamentoSNaPshot® para detecc¸ãode
cincomutac¸õesdoDNAmitocondrial:A1555GeA827Gnogene12SrRNAeA7445G,7472insC eT7511CnogenetRNASer(UCN).Paralelamente,utilizamosareac¸ãodepolimeraseemcadeiae
sequenciamososprodutosparatriagemdasmutac¸õesnogeneGJB2nospacientesportadores demutac¸õesnoDNAmitocondrial.
Resultados: Emnossapopulac¸ão,nãoconseguimosdetectarapresenc¸adamutac¸ãoA1555Gno gene12SrRNAenemasmutac¸õesA7445G,7472insCeT7511CnogenetRNASer(UCN).Entretanto,
constatamosqueseispacientes(3,37%)eramportadoresdamutac¸ãohomozigotaA827G;eum delestambémportavaamutac¸ãohomozigotaGJB2235delC.
Conclusão:Nossosachadosnopresenteestudoindicamque,mesmoempacientes esporádi-coscomdeficiênciaauditivasensorioneuralnãosindrômica,asmutac¸õesdoDNAmitocondrial tambémpodemcontribuirparaofenótipoclínico.
© 2015 Associac¸˜ao Brasileira de Otorrinolaringologia e Cirurgia C´ervico-Facial. Publicado por Elsevier Editora Ltda. Este ´e um artigo Open Access sob uma licenc¸a CC BY (http:// creativecommons.org/licenses/by/4.0/).
Introduction
Sensorineuralhearingloss (SNHL)is oneof themost
com-moncongenital disorders.The incidence is approximately
onein1000newbornsworldwide.1---3Overhalfofthemhave
ageneticcausewithautosomaldominant,autosomal
reces-sive,X-linkedormitochondrialpatternofinheritance.4,5Up
to now, more than 200 point mutations in mitochondrial
DNA(mtDNA) have been reportedin the mtDNA mutation
database MITOMAP.6 Among them,several mutations have
beenfoundtobeassociatedwithsyndromic,nonsyndromic
andaminoglycoside-induced hearing loss,especially those
in 12SrRNAand tRNASer(UCN) genes. The A1555G, which is
locatedinthehighlyconservedA-siteofmitochondrial12S
rRNA,wasthefirstonetobedescribedtobeassociatedwith
SNHLandfoundtoberesponsibleforboth
aminoglycoside-inducedand nonsyndromichearingloss inseveral families
worldwide.7---10 The A827G, a homoplasmic mutation in
mtDNA12SrRNA,wasrecentlyfoundtoberesponsiblefor
nonsyndromichearinglossinallmaternally relatedfamily
membersinaChinesefamily.11Italsohasbeenreportedto
beassociatedwithbothaminoglycoside-ototoxicityand
non-syndromichearingloss insporadic individuals.12,13 Besides
the12S rRNAgene, themitochondrial tRNASer(UCN) gene is
also associated with SNHL,as well as withnonsyndromic
deafness,asseveralmutationshavebeenidentified,
includ-ingA7445G,14,157472insC,16 T7511C.17,18
Although, asmentioned above,mtDNA mutations have
been reported to be associated with nonsyndromic SNHL
in severalfamilies,little is knownabout theincidence of
these mtDNA mutations in sporadic patients with
nonsyn-dromicSNHL.Ourprevious studywasfocusedontheGJB2
and SLC26A4 mutations in patients withautosomal
reces-sivenonsyndromichearingloss(ARNHL).Consequently,we
findthatalthoughtheGJB2andSLC26A4genearethemost
commondeafnessgenes,onlyasmallpartofthesepatients
carry homozygous or compound heterozygous pathogenic
mutations.19 Thatindicatesthatotherdeafnessgenesmay
contributetotheclinicalphenotype.Inthisstudyweuseda
rapidmethod,SNaPshot®sequencing,toscreen178sporadic
patients with nonsyndromic SNHL to estimate the
preva-lenceofmtDNAmutationsinsuchpopulation.Moreover,to
identify the role of the GJB2 gene in the deafness
phe-notype, we also screened the GJB2 gene in the patients
carryingmtDNAmutations.
Methods
Patientsandsamples
A total of 178 sporadic patients withnonsyndromic SNHL
participatedinthisinvestigation.Theyoriginatedfrom
var-iousregionsofourprovince.Eachpatientreceivedcareful
physicalexamination,andacomprehensiveclinicalhistory
wasrecorded.Patientswithdominantfamilyhistory,a
his-tory of aminoglycoside exposure, and syndromic hearing
loss were excluded from the study. All subjects
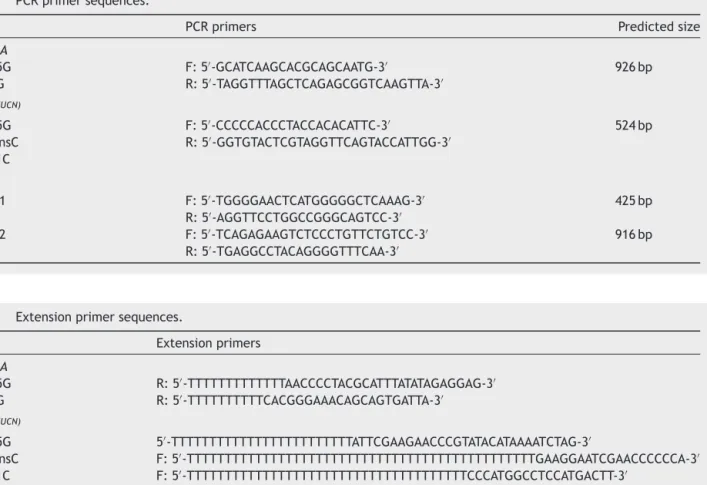
Table1 PCRprimersequences.
Gene PCRprimers Predictedsize
12SrRNA
A1555G F:5′-GCATCAAGCACGCAGCAATG-3′ 926bp
A827G R:5′-TAGGTTTAGCTCAGAGCGGTCAAGTTA-3′
tRNASer(UCN)
A7445G F:5′-CCCCCACCCTACCACACATTC-3′ 524bp
7472insC R:5′-GGTGTACTCGTAGGTTCAGTACCATTGG-3′
T7511C
GJB2
Exon1 F:5′-TGGGGAACTCATGGGGGCTCAAAG-3′ 425bp
R:5′-AGGTTCCTGGCCGGGCAGTCC-3′
Exon2 F:5′-TCAGAGAAGTCTCCCTGTTCTGTCC-3′ 916bp
R:5′-TGAGGCCTACAGGGGTTTCAA-3′
Table2 Extensionprimersequences.
Gene Extensionprimers
12SrRNA
A1555G R:5′-TTTTTTTTTTTTTAACCCCTACGCATTTATATAGAGGAG-3′
A827G R:5′-TTTTTTTTTTCACGGGAAACAGCAGTGATTA-3′
tRNASer(UCN)
A7445G 5′-TTTTTTTTTTTTTTTTTTTTTTTTATTCGAAGAACCCGTATACATAAAATCTAG-3′
7472insC F:5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGAAGGAATCGAACCCCCCA-3′
T7511C F:5′-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCCATGGCCTCCATGACTT-3′
response (ABR), otoacoustic emissions (OAE) and tympa-nometry.The severityof hearing impairmentwas defined asnormal(<26dB),mild (26---40dB),moderate(41---70dB), severe(71---90dB)andprofound (>90dB).Peripheralblood sampleswereobtainedfromthepatientsforgenomicDNA isolation.
Informedconsentwasobtainedfromadultpatientsand fromtheparentsofchildren,andthisstudywasapprovedby localEthicsCommittee(Ethical Committeeapproval num-ber:2013---007).
SNaPshot® sequencing
Afinal20LPCRreactionmixturecontained1×GCBuffer
I,3.0mMMg2+;0.3mMdNTPs,1UHotStarTaqpolymerase,
10ngtemplateDNA,and1Mprimermixture,includingtwo
pairsofprimerstoamplifyfivemutationregions(Table1).
ThePCRconditionswereasfollows:95◦Cfor2min;11cycles
of94◦Cfor20s,65---0.5◦Cpercyclefor40s,72◦Cfor90s;24
cyclesof94◦Cfor20s,59◦Cfor30s,72◦Cfor90s;72◦Cfor
2min.ForpurificationofPCRproducts,5Ushrimpalkaline
phosphatase(SAP)(Promega)and2UExonucleaseI
(Epicen-tre)wereaddedinto15LPCRproducts.Themixturewas
incubatedat37◦Cfor60min,followedbyincubationat75◦C
for15min.
Thesinglebaseextension(SBE)wasperformedinafinal
10Lreactionmixture,containing5LSNaPshot®Multiplex
Kit(AppliedBiosystems),2LpurifiedmultiplexPCR
prod-uct, and 0.8M extension primer mixture (Table 2). The
reactionprogramwas96◦Cfor1min;28cyclesof96◦Cfor
10s,52◦Cfor5s,60◦Cfor30s;4◦Cforever.Then,theSBE
products were purifiedby using SAP. For sequence
analy-sis,0.5LpurifiedmultiplexSBEproductsweremixedwith
0.5LLiz120SIZESTANDARD(AppliedBiosystems)and9L
Hi-Di(AppliedBiosystems),anddenaturedat95◦Cfor5min.
Then,theproductsweresequencedbytheABI3730XLDNA
sequencer.ThedatawasanalyzedbytheGeneMapperv4.1
software(AppliedBiosystems).
GJB2mutationdetectionbypolymerasechain reaction
Two exons of GJB2 were amplified from DNA samples by
polymerasechainreaction (PCR).The primerswere listed
in Table 1. PCR amplification and subsequent purification
wereperformed aswe describedbefore.ThepurifiedPCR
productsweresequencedusingthedideoxychainterminator
methodonan ABI 3730XL DNAsequencer(Applied
Biosys-tems)withtheABIBigDyeTerminatorv3.1CycleSequencing
Kit (Applied Biosystems), according tothe manufacturer’s
protocols.TheprimerswerethesameasthoseforthePCR
amplification.
Results
The studysamples consisted of82 males and96 females.
Table3 ClinicalandgeneticcharacteristicsofsixpatientscarryingA827GinmtDNA12SrRNA.
Patient Sex Ageofonset(years) Levelofhearingimpairment A827GinmtDNA12SrRNA MutationsinGJB2
Right Left
#61 Male 0 Profound Profound +/+ 235delC/235delC
#105 Female 0 Profound Profound +/+ WT/WT
#131 Female 0 Profound Profound +/+ WT/WT
#140 Female 0 Profound Profound +/+ WT/WT
#197 Female 0 Profound Profound +/+ WT/WT
#210 Male 0 Profound Profound +/+ WT/WT
years,with a mean age of 5.53±4.44 years.The results of audiometric testing revealed cochlear involvement in allparticipantsand thatallof them sufferedfromsevere (71---90dBtoprofound(>90dB))bilateralsensorineural hear-ingimpairment.
In all 178 sporadic patients with nonsyndromic SNHL, wefailedtodetectthepresenceoftheA7445G,7472insC, T7511CmutationsinthetRNASer(UCN)gene.Moreover,wedid
notdetect A1555G mutationin the 12SrRNAgene inthis population. However, the A827G variant in the 12S rRNA genewasfoundin 6patients,whoallcarriedhomozygous mutation.To illuminate the role of the GJB2 genein the phenotypicexpressionintheindividualswithmtDNA muta-tions,wescreenedtheGJB2genemutationsinthesubjects carryingA827Gmutation.Consequently, onlyone patient, whohadhomozygous235delC,carriedpathogenicmutation
intheGJB2gene(Table3).
Discussion
Most of the previously reported studies focused on the
association between the mtDNA mutations and the
pop-ulationwithaminoglycoside-induced and/ornonsyndromic
hearingloss.12,19SeveralmtDNAmutations,suchasA1555G
and A827G in the 12S rRNA gene, A7445G, 7472insC and
T7511CinthetRNASer(UCN)genehave beenidentifiedtobe
associatedwithsuchpopulations.10,13,14,19,20However,these
mutationsare often notsufficient to producethe clinical
phenotype.14,19 Inpatients withARNHL,similarwithother
studies,ourpreviousstudyfoundthatabout30---40%patients
carriedpathogenicmutationsinthemostcommondeafness
genes,butnearly40%ofthesepatientscarriedheterozygous
mutation.Thus, other genemutations may together
con-tributetothephenotype.Althoughtheassociationbetween
the mtDNA mutations and nonsyndromic hearing loss has
beenidentifiedinseveralfamilies,littleisknownaboutthe
prevalence of mtDNA mutations in sporadic patients with
nonsyndromicSNHL.
Mitochondrial DNA mutations in tRNA genes can cause
tRNAmodificationand afailurein tRNAmetabolism, thus
impairingtheproteinsynthesis andreducing theATP
syn-thesis, which are considered to be the main pathogenic
factors.21---25 TheA7445G mutationwasfirstidentifiedina
familywithnonsyndromicdeafness.26Thismutationleadsto
the3′endendonucleolyticprocessingdefectintheL-strand
polycistronic RNA precursor.27 Consequently, the A7445G
mutation can cause a reduction of more than 50% in the
tRNASer(UCN)levelandadecreaseinproteinsynthesis,
play-ingadeterminantroleintherespiratoryphenotypeofthe
mutantcelllines.28,29 The7472insCwasoriginallyreported
byTirantietal.,whichislikelytoalterthestructureofthe
TpsiCloopinthetRNASer(UCN)cloverleafsecondary
struc-ture,andhasbeenprovedtoimpairboth5′and3′processing
of the tRNASer(UCN) and cause a drop in the steady-state
level of the tRNASer(UCN).16,24,30 The T7511C mutation has
beenidentifiedtobeassociatedwithnonsyndromichearing
loss in several differentethnic families.17,18,31 This
muta-tion can affect the processing of L-strandRNA precursor,
spanning tRNASer(UCN) aswell asND6 mRNA, specifically in
the 5′ end of tRNA.23,32 As a result, theT7511C mutation
causesareductionoftheleveloftRNASer(UCN)andan
impair-mentinmitochondrialproteinsynthesis.22,28Althoughthese
mutationsinthetRNASer(UCN)havebeenfoundtobe
associ-atedwiththepatientswithaminoglycoside-inducedand/or
nonsyndromic hearing loss, many studies failed todetect
such mutations in those patients. In the study reported
by Xing etal., theyanalyzed the molecular
characteriza-tionof a Chinesefamilywithaminoglycoside-induced and
nonsyndromic hearing loss, but did not find the A7445G,
7472insC,andT7511CmutationsinthetRNASer(UCN) gene.33
InanArgentineanfamilywithaminoglycoside-induced
hear-ing loss,such mutations werealsonot found.34 Moreover,
Liet al. performed amolecular analysis in 164unrelated
Caucasian individuals with nonsyndromic hearing
impair-ment and failed to detect the presence of the A7445G,
7472insC,andT7511CmutationsinthetRNASer(UCN) gene.12
Abreu-Silva et al. also did not find these mutations in
203unselectedBrazilianhearing-impairedpatients.35Upto
now, little is known about the incidence of these
muta-tions in the tRNASer(UCN) gene in sporadic patients with
nonsyndromic SNHL. In the present study, we explored
thesemutationsinsuchpopulation,butnoneofthese
sub-jectswasfoundtocarrysuchmutationsinthetRNASer(UCN)
gene.
The A1555G mutation creates a specific GC base pair,
whichmakesthesecondarystructureofmitochondrial12S
rRNAmoreclosely resemblingthecorrespondingregionof
E. coli 16S rRNA and binding aminoglycosides with high
affinities.36 Thus, the patients carrying this mutation can
suffer from SNHLafter using aminoglycosides.13 However,
previous studies have identified that A1555G mutation in
12S rRNA is also associated with nonsyndromic hearing
muchlowerthaninaminoglycoside-inducedhearing
impair-ment.Luetal.reportedthattheincidencesoftheA1555G
mutation were 1.43% and 10.41% in a Chinese pediatric
populationwithnonsyndromicandaminoglycoside-induced
hearing loss, respectively.19 Similarly, in a Polish
popula-tion,Rydzaniczetal.observedthattheincidencewas5.5%
and 1.6% in the cohorts withaminoglycoside-induced and
nonsyndromichearingloss.37 InaCaucasianpediatric
non-syndromic hearing loss population, the frequency of the
A1555G mutationwas0.6%.12 In thisstudy,we found that
none of the subjects carriedthe A1555G mutation.
Com-paredwithprevious reports, theincidenceof theA1555G
mutationinourpopulationwithnonsyndromicSNHLis
rela-tivelylower.
The A827G mutation,like A1555G mutation,is located
at the A-site of the mitochondrial 12S rRNAgene, which
is highlyevolutionarilyconservedindifferent species.34 It
is possible that the alteration of the tertiary or
quater-nary structure of the 12S rRNA by the A827G mutation
may lead tomitochondrial dysfunction;thereby, it would
play a role in the pathogenesis of hearing loss.11,34 The
pathogenesisofthismutationhasbeenidentifiedina
Chi-nesefamilywithnonsyndromichearingloss.11Althoughthe
A827Gmutationwasalsofoundinnormalhearingcontrols,19
morestudiesconsideritasapathogenicmutation
depend-ing on its location and absence in the controls in their
studies.11---13,34,37Meanwhile,thesestudiesalsofound
incom-plete penetrance of the A827Gmutation,which indicates
thatthismutationaloneisnotsufficienttoproduceclinical
phenotype.11,34Thus,somenormalhearingsubjectscarrying
theA827Gmutationmaybeattributedtoincomplete
pen-etrance.Inourstudy,wefoundthat3.37%(6/178)patients
carryhomozygousA827Gmutation.InanotherChinese
spo-radicpediatricpopulationwithnonsyndromichearingloss,
theincidenceis4.41%.13TheincidenceoftheA827G
muta-tion in such Chinese population seems to be higher than
inotherethnicgroups.11,37Aspreviousstudiesshowedthat
expressionoftheclinicalphenotypeofdeafness-associated
homoplasmicmutationsinthe12SrRNAgenerequires the
contribution of modulatingfactors, including
aminoglyco-sides or nuclear modifier genes, and the GJB2 gene is
a potential candidatemodifier gene,12,38---40 we performed
mutationalscreeningoftheGJB2geneinthepatients
car-rying the A827G mutation. As a result, only one patient
carried both 12S rRNA A827G mutation and homozygous
GJB2235delCmutation.ThelackofGJB2mutationinother
patients indicates that other nuclear modifier genes and
otherenvironmentalfactorsmaycontributetotheclinical
phenotypeinthesepatients.
Conclusion
InthepresentstudywescreenedmtDNAmutationsin
spo-radic patients withnonsyndromic SNHL and found a total
of 6 subjects (3.37%) carrying homozygous mtDNA
muta-tions.Ourfindingsindicatethatevensporadicpatientswith
nonsyndromicSNHLalsocarrymtDNAmutationsandthese
mutationsmightcontributetotheclinicalphenotype.
Fur-therstudiesscreeningthemutationsinmtDNAareneeded
to help us understand the prevalence and role of mtDNA
mutationsinsporadicpatientswithnonsyndromicSNHL.
Conflicts
of
interest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgements
This work was supported by grants from the Department
ofPublicHealth(no.2011ZDA012)andScienceTechnology
Department(no.2014C37036).
References
1.MortonNE.Geneticepidemiologyofhearingimpairment.Ann NYAcadSci.1991;630:16---31.
2.WangQJ,ZhaoYL,RaoSQ,GuoYF,He Y,LanL, etal. New-bornhearingconcurrent genescreeningcanimprovecarefor hearingloss:astudyon14,913Chinesenewborns.IntJPediatr Otorhinolaryngol.2011;75:535---42.
3.DaiP,LiuX,YuF,ZhuQ,YuanY,YangS,etal.Molecular etiol-ogyofpatientswithnonsyndromichearinglossfromdeaf-mute schoolsin18provincesofChina.ZhonghuaErKeXueZaZhi. 2006;4:1---5.
4.BayazitYA,YilmazM.Anoverviewofhereditaryhearingloss. ORLJOtorhinolaryngolRelatSpec.2006;68:57---63.
5.MortonCC.Genetics,genomicsandgenediscoveryinthe audi-torysystem.HumMolGenet.2002;11:1229---40.
6.The mitochondrial DNA mutation database. http://www.mitomap.org/[accessed20.07.14].
7.Fischel-GhodsianN,PrezantTR, BuX,OztasS.Mitochondrial ribosomalRNAgenemutationinapatientwithsporadic amino-glycosideototoxicity.AmJOtolaryngol.1993;14:399---403. 8.PandyaA,XiaX,RadnaabazarJ,BatsuuriJ,DangaansurenB,
Fischel-GhodsianN, etal. Mutationinthemitochondrial 12S rRNAgeneintwofamiliesfromMongoliawithmatrilineal amino-glycosideototoxicity.JMedGenet.1997;34:169---72.
9.delCastilloFJ,Rodríguez-BallesterosM,MartínY,ArellanoB, Gallo-TeránJ,Morales-AnguloC,etal.Heteroplasmyforthe 1555A>G mutationinthemitochondrial 12SrRNAgeneinsix Spanishfamilieswithnon-syndromichearingloss.JMedGenet. 2003;40:632---6.
10.LuJ, QianY, Li Z,YangA, ZhuY, Li R, et al.Mitochondrial haplotypesmaymodulatethephenotypicmanifestationofthe deafness-associated 12S rRNA 1555A>G mutation. Mitochon-drion.2010;10:69---81.
11.XingG,ChenZ,WeiQ,TianH,LiX,ZhouA,etal.Maternally inheritednon-syndromichearinglossassociatedwith mitochon-drial12SrRNAA827G mutationina Chinesefamily. Biochem BiophysResCommun.2006;344:1253---7.
12.Li R, GreinwaldJHJr, YangL, ChooDI, Wenstrup RJ, Guan MX. Molecular analysis of the mitochondrial 12S rRNA and tRNASer(UCN)genesinpaediatricsubjectswithnon-syndromic hearingloss.JMedGenet.2004;41:615---20.
13.LiZ,LiR,ChenJ,LiaoZ,ZhuY,QianY,etal.Mutationalanalysis ofthemitochondrial12SrRNAgeneinChinesepediatric sub-jectswithaminoglycoside-inducedandnon-syndromichearing loss.HumGenet.2005;117:9---15.
14.Fischel-GhodsianN,PrezantTR,FournierP,StewartIA,MawM. Mitochondrialmutationassociatedwithnonsyndromicdeafness. AmJOtolaryngol.1995;16:403---8.
15.Hutchin TP, LenchNJ, Arbuzova S, Markham AF,Mueller RF. Maternally inherited hearing impairment in a family with the mitochondrial DNAA7445G mutation. EurJHum Genet. 2001;9:56---8.
associated with a novel point mutation in mitochondrial tRNASer(UCN)gene.HumMolGenet.1995;4:1421---7.
17.SueCM,TanjiK,HadjigeorgiouG,AndreuAL,NishinoI,Krishna S, et al. Maternally inherited hearing loss in a large kin-dredwithanovelT7511CmutationinthemitochondrialDNA tRNA(Ser(UCN))gene.Neurology.1999;52:1905---8.
18.IshikawaK,TamagawaY,TakahashiK,KimuraH,KusakariJ,Hara A,etal.Nonsyndromichearinglosscausedbyamitochondrial T7511Cmutation.Laryngoscope.2002;112:1494---9.
19.LuJ,LiZ,ZhuY,YangA,LiR,ZhengJ,etal.Mitochondrial 12SrRNAvariantsin1642HanChinesepediatricsubjectswith aminoglycoside-inducedand nonsyndromichearingloss. Mito-chondrion.2010;10:380---90.
20.DingY,LengJ,FanF,XiaB,XuP.TheroleofmitochondrialDNA mutationsinhearingloss.BiochemGenet.2013;51:588---602. 21.McKenzieM,LiolitsaD,HannaMG.Mitochondrialdisease:
muta-tionsandmechanisms.NeurochemRes.2004;29:589---600. 22.Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA,
Guan MX. Biochemicalcharacterization ofthe mitochondrial tRNASer(UCN)T7511Cmutationassociatedwithnonsyndromic deafness.NucleicAcidsRes.2004;32:867---77.
23.Li X, Zhang LS, Fischel-Ghodsian N, Guan MX. Biochemi-calcharacterizationofthedeafness-associatedmitochondrial tRNASer(UCN)A7445Gmutationinosteosarcomacellcybrids. BiochemBiophysResCommun.2005;328:491---8.
24.Toompuu M, Levinger LL, Nadal A, Gomez J, Jacobs HT. The 7472 insC mtDNA mutation impairs 5′ and 3′
processingoftRNA(Ser(UCN)).BiochemBiophysResCommun. 2004;322:803---13.
25.TangX,LiR,ZhengJ,CaiQ,ZhangT,GongS,etal.Maternally inheritedhearinglossisassociatedwiththenovelmitochondrial tRNASer(UCN)7505T>CmutationinaHanChinesefamily.Mol GenetMetab.2010;100:57---64.
26.ReidFM,VernhamGA,JacobsHT.Anovelmitochondrialpoint mutationinamaternalpedigreewithsensorineuraldeafness. HumMutat.1994;3:243---7.
27.LevingerL,JacobsO,JamesM.Invitro3′-endendonucleolytic
processing defect in a human mitochondrial tRNA(Ser(UCN)) precursor with the U7445C substitution, which causes non-syndromicdeafness.NucleicAcidsRes.2001;29:4334---40. 28.Guan MX,Enriquez JA, Fischel-Ghodsian N,Puranam RS,Lin
CP,MawMA,etal.Thedeafness-associatedmitochondrialDNA mutationatposition7445,whichaffectstRNASer(UCN) precur-sorprocessing,haslong-rangeeffectsonNADHdehydrogenase subunitND6geneexpression.MolCellBiol.1998;18:5868---79. 29.ReidFM,RovioA,HoltIJ,JacobsHT.Molecularphenotypeof
ahumanlymphoblastoidcell-linehomoplasmicforthenp7445
deafness-associatedmitochondrialmutation.HumMolGenet. 1997;6:443---9.
30.ToompuuM,TirantiV,ZevianiM,JacobsHT.Molecular pheno-typeofthenp7472deafness-associatedmitochondrialmutation inosteosarcomacellcybrids.HumMolGenet.1999;8:2275---83. 31.Chapiro E,Feldmann D,Denoyelle F,Sternberg D,Jardel C, EliotMM,etal.TwolargeFrenchpedigreeswithnonsyndromic sensorineural deafness and the mitochondrial DNA T7511C mutation:evidenceforamodulatoryfactor.EurJHumGenet. 2002;10:851---6.
32.Puranam RS, Attardi G. The RNase P associated with HeLa cellmitochondriacontainsanessentialRNAcomponent iden-ticalinsequencetothatofthenuclearRNaseP.MolCellBiol. 2001;21:548---61.
33.XingG, Chen Z, Wei Q, TianH, Li X, Zhou A, et al. Mito-chondrial12SrRNAA827Gmutationisinvolvedinthegenetic susceptibilitytoaminoglycosideototoxicity. BiochemBiophys ResCommun.2006;346:1131---5.
34.ChaigMR,ZernottiME,SoriaNW,RomeroOF,RomeroMF,Gerez NM.Amutationinmitochondrial 12SrRNA,A827G,in Argen-tineanfamilywithhearinglossafteraminoglycosidetreatment. BiochemBiophysResCommun.2008;368:631---6.
35.Abreu-SilvaRS, Lezirovitz K,Braga MC, SpinelliM, Pirana S, Della-Rosa VA, et al. Prevalence of the A1555G (12S rRNA) andtRNASer(UCN)mitochondrialmutationsinhearing-impaired Brazilianpatients.BrazJMedBiolRes.2006;39:219---26. 36.Hamasaki K, Rando RR. Specific binding of aminoglycosides
to a human rRNA construct based on a DNA polymorphism whichcausesaminoglycoside-induceddeafness.Biochemistry. 1997;36:12323---8.
37.Rydzanicz M, Wróbel M, Pollak A, Gawecki W, Brauze D, Kostrzewska-PoczekajM,etal.Mutationanalysisof mitochon-drial 12S rRNA gene in Polish patients with non-syndromic andaminoglycoside-inducedhearingloss.BiochemBiophysRes Commun.2010;395:116---21.
38.Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ,etal. Mitochondrialribosomal RNAmutationassociated withbothantibiotic-inducedandnon-syndromicdeafness.Nat Genet.1993;4:289---94.
39.Li R,Xing G, YanM,CaoX, LiuXZ,Bu X, etal. Cosegrega-tionofC-insertionatposition961withtheA1555Gmutation ofthemitochondrial 12S rRNAgenein alarge Chinese fam-ilywithmaternallyinheritedhearingloss.AmJMedGenetA. 2004;124A:113---7.