ORIGINAL
ARTICLE
Signal to cut-off (S/CO) ratio and detection of HCV genotype 1 by
real-time PCR one-step method: is there any direct relationship?
Authors
GuilhermeAlbertoni1
CPArnoni1
PRBAraújo1
FOCarvalho1
JABarreto1
1Colsan–Associação BeneficentedeColetade Sangue,SãoPaulo,SP, Brazil
Submittedon:05/12/2009 Approvedon:06/07/2009
Correspondence to:
GuilhermeAlbertoni Av.Jandira,1260, Indianópolis SãoPaulo–SP–Brazil CEP:04080-006 Phone:551150556588/ R.39
Fax:551150556588 E-mail:albertonig@nefro. epm.br
Wedeclarenoconflictof interest.
ABSTRACT
Background: Polymerase chain reaction (PCR) methods play an essential role in providing data relatedtodiagnosis,monitoringandtreatmentofhepatitisCvirus(HCV)infection.EIAresultsare reportedas“reactive”or“nonreactive”andEIAS/COratiomayalsobereportedas“high”or“low.” Thisstudyaimedtoevaluatetheperformanceofareal-timeRT-PCRandassesswhetherthereis relationshipbetweenS/COandPCRresults.Study Design and Methods: Serafromblooddonors wereanalyzedbyEnzyme-LinkedImmunosorbentAssay(ELISA)andRT-PCRassaytodetectHCV infection.Results: TheRT-PCRassaytogenotypes1a/bshowedanacceptablelinearresponseinse-rialdilutions.Thesamplesweredividedintotwogroupsbasedontheirserologicalresults:groupA– S/COratio<3(60samples)andgroupB–S/COratio>3(41samples).Viralloadswereconfirmed positiveingroupBsamplesin90%,andingroupAsampleswereconfirmedpositiveinonly13%by RT-PCR.Conclusion: ThemethodologyusedwasabletodetectthepresenceofRNA-HCVgenotype Iin90%ofthesamplesserologicallypositiveingroupB.Allnegativesamplesweresenttosearchfor othergenotypesofHCV(genotypes2-6)andwereconfirmedasnegative.Thesedatasuggeststhat thesenegativesamplesmayhaveHCVRNAviralloadbelowthedetectionlimitofourtest(310IU/ mL),orafalsepositiveresultinserologicaltest,orspontaneousviralclearanceoccurred.
Keywords:HCV,HCVgenotype1,real-timePCR,ELISA,RNAextraction.
[BrazJInfectDis2010;14(2):147-152]©ElsevierEditoraLtda.
INTRODUCTION
The hepatitis C virus (HCV), discovered in 1989,isthemajorcausativeagentofhepatitis non-A,non-B.Thisvirus,whichbelongstothe familyFlaviviridae andhasagenomeconstitut- edbyapositivesingle-strandRNA,istransmit-ted parenterally, showing a high incidence in hemophiliacs hemodialysis patients, injecting drugusers,andpatientswithpost-transfusion hepatitis.1,2
HepatitisCinfectionhashadhighpreva-lence in Brazilian population mainly in the southregion.3,4Accordingtoliterature,5more than half of infected patients (about 70 to 85%)developchronichepatitis,someofthem (10% to 20%) progressing to cirrhosis and hepatocellular carcinoma in a period of 10 to15years.BecausehepatitisChashighpo-tential to become chronic, it is considered a seriouspublichealthproblem,beingestimat-edthatthereare170millionpeopleinfected worldwide.6 Currently,hepatitisCisthelead-ingcauseoflivertransplantsinUSA.7
Another molecular technique that has been used to differentiateHCVsubtypesisgenotyping.Themolecular characterizationofgenotypesandsub-genotypesofHCV isparticularlyimportantinguidingtheresponsetotreat-mentanddiseaseprognosis.5,6,14 Thereare11majorgeno- typesofHCVand23subtypes.Theprevalenceofgeno-types of HCV varies according to geographic region. In Brazil,thegenotypes1,2,and3arethemostoftenfound.6 Accordingtoliterature,patientsinfectedwithgenotypes 2and3respondbettertotreatmentthanthoseinfected withgenotype1.Genotype1patientshavealesseffective responsetotreatmentandahighviralload.15,16Hepatitis C treatment is based on therapy with interferon-α and Ribavirin.Itisaverylongtreatmentwithanaveragedu-rationof48to72weeks,17withahighcost(betweenU$ 12,000andU$15,000U.S.dollarsperpatientperyearin theU.S).Besidesbeinganexpensiveandprolongedtreat-ment,ithasasuccessfulrateof45-80%dependingonviral genotype,17,18probablybecauseonlyfewHCVgenotypes respondquickandcompletelytothistherapy.Thefactis that there is a strong relationship between genotype of thevirusandeffectivenessoftreatmentincreasetheim-portanceofgenotypingtheHCV.Genotypingassessthe patient’sprognosisprovidingimportantinformation,so thatthecliniciancantailorthetreatmenttothecase.
PCR
PCRdependsontheabilitytoalternatelydenature(melt) double-stranded DNA molecules and renature (anneal) complementarysinglestrandsinacontrolledfashion.The secondrequirementforPCRistheuseofsyntheticoligo-nucleotidesatleast18–20 nucleotideslongwithadefinedse-quence.Suchsyntheticnucleotidescanbereadilyproduced withautomatedinstrumentsbasedonthestandardreac- tionscheme.AtypicalPCRprocedurebeginsbyheat-de-naturationofaDNAsampleintosinglestrands.Next,two syntheticoligonucleotidescomplementarytothe3endsof thetargetDNAsegmentofinterestareaddedingreatex-cesstothedenaturedDNA,andthetemperatureislowered to50-60°C.Thesespecificoligonucleotides,whichareata veryhighconcentration,willhybridizewiththeircomple-mentarysequencesintheDNAsample,whereasthelong strandsofthesampleDNAremainapartbecauseoftheir low concentration. The hybridized oligonucleotides then serveasprimersforDNAchainsynthesisinthepresence ofdeoxynucleotides(dNTPs)andatemperature-resistant DNApolymerase,suchasthatfromThermus aquaticus(a bacteriathatlivesinhotsprings).Thisenzyme,calledTaq polymerase, can remain active even after being heated to 95° C and can extend the primers at temperatures up to 72° C. When synthesis is complete, the whole mixture is thenheatedto95°CtomeltthenewlyformedDNAdu-plexes.Aftertemperatureisloweredagain,anothercycleof
synthesistakesplacebecauseexcessprimerisstillpresent. Repeatedcyclesofmelting(heating)andsynthesis(cool-ing)quicklyamplifythesequenceofinterest.Ateachcycle, the number of sequence copies between the primer sites isdoubled;therefore,thedesiredsequenceincreasesexpo-nentially—aboutamillion-foldafter20cycles—whereas allothersequencesintheoriginalDNAsampleremainun-amplified.
Use of the PCR technique
The development of DNA segments amplification using PCRopenedvastprospectsforanalysisofgenes,diagnosis ofgeneticdiseases,anddetectionofinfectiousagents,such as cytomegalovirus, hepatitis B and C viruses, herpes vi-russimplex,rubellavirus,humanimmunodeficiencyvirus (HIV),Chlamydia trachomatis, Helicobacter pylori, Pneu-mocystis jirovecii(formerlyP.carinii),andMycobacterium tuberculosis.
The advancement of science on the understanding of genes brought into the laboratory routine genetics tools forthediagnosisinmolecularlevel.Inthelineofgenetic diseases,thecontinuingdevelopmentofnewprotocolshas enabledthedetectionofsmallchangesinDNAsequence; forexample,incysticfibrosis.
OtherapplicationsespeciallyforthePCRistheclon-ingofaDNAspecificfragment,whichmaybeageneand knowledgeofcodingDNA(cDNA)obtainedfromthemol-eculeofRNA,whichallowsthestudyofgeneexpression.
Finally, PCR has great potential in forensic medicine. Itssensitivitymakesitpossibletouseaverysmallsample (minimumtracesofbloodandtissuesthatcouldcontain theremainsofonlyonecell)andstillgeta“DNAfinger-print”ofthepersonfromwhichthesamplewascollected andcanthereforemakecomparisonswiththoseobtained fromvictimsand/oracriminalsuspect.19
Real-time PCR
Theabilitytomonitorreal-timePCRhasrevolutionizedthe process of DNA and RNA fragment quantification. Real-time PCR makes nucleic acid quantification with greater accuracyandreproducibility,aswellasitspredictivevalues duringtheexponentialphaseofthereaction.Thepointthat detectsthecycleinwhichthereactionreachesthethreshold ofexponentialphaseiscalledCycleThreshold(CT).Inreal-timePCR,eachroundofamplificationleadstotheemission ofafluorescentsignalandthenumberofsignalspercycle isproportionaltotheamountofHCV-RNAinthestarting sample.20 Thisallowstheaccurateandreproduciblequanti-ficationbasedonfluorescence.
ingeverycycleandrepresenttheamountofproductampli-fied.ThemostusedcompoundsarefluorescentSYBRGreen andTaqManprobe.
Real-time PCR requires an instrumentation platform thatconsistsofathermalcycler,computer,opticsforfluo- rescenceexcitationandemissioncollection,anddataacqui-sitionandanalysissoftware.
Theaimofthisworkistoevaluatereal-timePCROne-StepmethodtodetecthepatitisCvirusgenotype1andtry toestablisharelationshipbetweenS/COratioandPCRtest results.
MATERIAL AND METHODS
Clinical specimens
FromOctober2008toAugust2009,bloodsampleswereob- tainedfrom101blooddonorswithpositiveantibodyscreen-ing for hepatitis C (ELISA – EIA-3; Biomeriéux Co.) and 10donorswithnegativescreening.Allbloodsampleswere negativeforothertestsmandatoryinbloodbankscreening (ELISA).
Design of primers and probe (FAM – Assay with fl uorescent marker/MGB – Minor Groove Binder)
Theprimersandprobeforreal-timePCRusedinthisstudy weredesignedinprimerexpresssoftware(AppliedBiosys-tems) based on HCV sequence. The sequences of primers HCVarethefollowing:5´-CGGGAGAGCCATAGTGGT–3’ (HCV1F,position:130-147);5´-CGCGACCCAACACTACTC –3´(HCV1R,position:256-273).Theprobesequencewas selectedwithinprimerpairHCV1FandHCV1Randcit-edatposition149-169,whichwasdesignedtobeperfectly complementarytothetargetsequencein5´-NCRofHCV genome.Thefollowingistheprobesequence:FAM–TGCG- GAACCGGTGAGTACACC–MGB(HCV1P).Bothprim-ersandprobeweresuspendedinDEPCWaterandstoredat -20°C.
RNA extraction
ForpreparationoftotalRNAfromhumansera,weusedthe QIAampViralRNAkitaccordingtomanufacturer’sinstruc-tions(QIAGEN,Courtaboeuf,France).Inbrief,140µLof serumwasincubatedwith560µLoflysisbuffercontaining chaotropicsaltsandcarrierRNAfor10minatroomtem- perature.Aftertheadditionof560µLofethanol,theprecip-itatedRNAwasappliedontoasilica-basedspincolumnfor purificationandwasfinallyelutedwith60µLofQIAamp elutionbuffer.
Reverse transcription and amplifi cation
Thereversetranscriptionwasperformedusingamethodol-ogyone-stepwithTaqManProbesReactionMix,containing 0,5µLSuperscriptIIIRT/PlatinumTaqMix;12,5µL(2xRe-actionMix);0,5µLForwardprimer(10uM);0,5µLReverse
primer(10µM);0,1µLFluorogenicprobe(10uM);0,5µL RNaseOut;<10µLTemplate(1pgto1µgtotalRNA)and DEPC-treatedwaterto20µL.
Amplificationwasperformedin20µLreactionmixture containing TaqMan Universal PCR Master Mix, UNG, DNTPs, a passive reference (6-carboxy-rhodamine; ROX). Standard cycling program was: 15 min hold at 50°CforcDNAsynthesistemperaturemayrangefrom 42-60°C;2minholdat95°C;40cyclesof:15sat95° C;30sat60°C.
Enzyme Linked Immunosorbent Assay Screening (ELISA)
TestingforHCVantibodieswasperformedwithathird generation enzyme immunoassay (EIA-3; Biomeriéux Co.) following the manufacturer’s instructions. S/CO ratio(S=sampleratioandCO=Cutoffratio)wasde-termined for all blood samples and, based on this, we createdtwogroups:groupAinwhichS/COratiowas< 3,andgroupBinwhichS/COratiowas>3.Thisalgo- rithmwasdevelopedinordertoestablishtherelation-ship between S/CO ratio and the probability RT-PCR beenpositive.21
RESULTS
Linear range and limit of detection
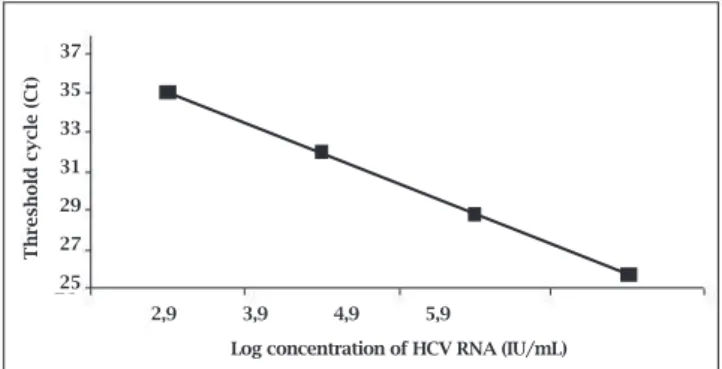
Oncethestandardcurvehadbeencalculated,thesoftware automaticallyquantifiedthenumberofHCVRNA,report-ingtheCtofthesampletotheCtofthestandardcurve.The correlation coefficient (R2) of the analysis was above 0.99 foralltheexperiments,fromacommercialstandardsample (positivecontrol)withapre-establishednumberofcopies. The standard curve was made using different concentra-tions(rangingfrom310to310.000IU/mLor2,5to5,5Log) showedinFigure1.
Figure 1: Standard curve of the HCV RNA concentration (log)
in serial dilutions vs. cycle number (Ct). Each dot represents
the results of triplicate PCR amplification for each dilution.
Log concentration of HCV RNA (IU/mL)
Threshold cycle (Ct)
2,9 3,9 4,9 5,9 37
35
33
31
29
27
DISCUSSION
Althoughtheriskoftransfusion-transmittedHCVhaspro-gressivelydecreasedduringthepastfewdecades,thanksto introductionofseveralpreventivemeasuressuchascareful selectionofblooddonors,enhanceddonorquestioningfor eligibilityandrefinementinblooddonationscreening,and stimulation to donor fidelity, the implementation of NAT togetherwiththeexistingserologicalassayscanreducethe residualriskofrecipient’sinfectionbyshorteningthewin- dowperiod(temporalgapspanningfromthetimeofinfec- tiontoseroconversion).Duringthisperiod,aninfecteddo-normayharborlargeamountsofinfectionviralparticlesin absenceofserologicalmarkersand/orsignsandsymptoms ofanongoinginfection.Duetotherelativelylongseronega- tivewindowperiodforHCVinfection,about82daysifus-ingthethirdgenerationantibodytestavailable,inthisstudy weshowedtheimportanceoftestingforviralRNA,which enables detection 10 days after infection and leads to safe transfusions.
Polymerase chain reaction (PCR) is a powerful tool fordetectionofminimalamountsofnucleicacids.Due totheexponentialamplificationofthetargetsequence,it hasexquisitesensitivity.Fewcopiesofanytranscriptcan be readily detected, even against a high background of unrelatednucleicacid.TheestablishmentofPCR-based detection methods has provided the basis for rapid and reliabledetectionofviralnucleicacidsintheclinicalset- ting.Itwasshownthatmonitoringvirusloadandkinet-ics of virus proliferation have prognostic relevance for thecourseofdiseaseandclinicaloutcome.22-27 Theavail-abilityofquantitativevirusdetectiontestsisthereforeof paramount importance for the clinical management of virusinfections.
MostassaysdescribedforthedetectionofviralDNAor RNAisbasedontheuseofhydrolysisprobes,alsoreferred toasTaqMan.Theprobesbindtothetargetstrandbetween PCRprimers.Theyareduallylabeledwithafluorescencere-porterdyeattachedtothe5´-end,aquencherdye,attached to the 3´-end. When the reporter molecule on TaqMan probeisstimulated,anappropriatelightsourcetoemitfluo-rescence,theenergyistransferredtothequencher,thereby suppressingtheemissionoffluorescencebythereporter.28 During PCR, when DNA polymerase extends the primers, thehybridizedprobesaresplitbytheenzyme5´exonuclease activityandthecorrespondingquencherandreportermol-eculesareseparated.Theenergytransferredtothequencher moleculeisthusabrogated,andthereporterstartsemitting fluorescence,whichcanbemeasuredattheendofeachex-tensionstep.29 Inwellestablishedassays,thereisalinearcor-relationbetweenthenumberofreleasedreportermolecules andthenumberofampliconssynthesizedduringeachPCR cycle.Thiscorrelationservesasabasisforcalculationofini-tialcopynumberoftargettranscript.
Figure 4: Samples with S/CO ratio >3.
Inordertocomparethereproducibilitybetweenourtest and a commercial test, we studied 24 samples by another PCR method (Amplicor Monitor HCV RNA assay®, Ver-sion 2.0, Roche Diagnostics, Meylan, France). All positive sampleswereconfirmedpositiveinbothmethods,showing 100%reproducibility(Figure2).
IngroupAsamples,fromatotalof60samples,onlyeight (13%)werepositivebyreal-timePCRtest(Figure3).
Outof41positivesampleswithS/COratio>3,37sam-ples(90%)wereconfirmedpositivebyRT-PCRtestingroup B,whilefoursampleswerenegative(Figure4).
Figure 3: Samples with S/CO ratio < 3.
Figure 2: Comparison between Real Time RT-PCR vs.
Amplicor Roche.
Comparison between Real-Time PCR vs. Amplicor.
Roche
25
20
15
10
5
0
Real Time PCR (positive samples) Amplicor/Roche (positive samples) Real Time PCR (negative samples) Amplicor/Roche (negative samples)
S/CO ratio < 3 and RT-PCR detection
13%
87%
RT-PCR (+) RT-PCR (-)
S/CO ratio < 3 and RT-PCR detection
10%
90%
Thismethodshowsseveraladvantages,suchasfastex-ecutionandreducedriskofcontamination,duetothelack ofpost-PCRprocessingsteps.TaqMantechnologyrequires specificconditionsforthechoiceofprimersandprobe:They shouldhaveaTmdifferenceof10°C,theabsenceofgua-nosineatthe5´aminoprobesequenceandanamplification lengthideallyunder100bp.Theampliconlength,together withtherestrictiveconditionsofprimersandprobedesigns, doesnotfacilitatetheassayusewithRNAviruses,inparticu-lar,duetotheirgeneticvariability.ThedevelopmentofDNA probeswithconjugatedminorgroovebinder(MGB)groups hasallowedtheuseofshorterprobesinhybridization-based assays.Interestingly,thedesignofanarraywithintherare conservedregionof5´-UTRofHCVwasfacilitatedbythe useofa3´-MGBprobe.
Inourlaboratory,theTaqManquantitativeHCVassay showedlinearityoverarangeof3,1x102to3,1x105IU/mL. Thus,serumsampleswithaviralloadhigherthan310.000 IU/mL have to be diluted. The detection limit of this test canvaryasnotedinotherpublicationsthatshowedanesti-matedlimitat550IU/mL30,31becausedifferentmethodsare usedfordeterminingRNAconcentration.32-35
Traniet al.36 also used the technique of one-step real-timePCRwithMGBprobetodetectavianinfluenzaviruses. Thelinearcorrelation(R2)ofthatstudywas0.99,thesame foundinourstudy,showinghighsensitivityoftest.Arruda
et al.,37 evenusingadifferenttechnology,SYBRGreentech-nique,toevaluatepatientswithHTLVI/IIinfection,madea standardcurveusingserialdilutionsofplasmidinconcen-trationsfrom100to105,gettingR2=0,89.
WangLu-Nanet al.,38throughtechnologyRocheCobas Amplicormethod,evaluatedR2fordetectionofHCVand wasabletodetectarangeof600to850,000IU/mL,unlike thedatapresentedbyus,demonstratingthatthetechnique usedbyourlaboratorywasmoresensitiveindetectinglow levelsofviralload.
EIA results are reported as “reactive” or “nonreactive” andEIAsignaltocutoff(S/CO)ratiomayalsobereported as“high” or“low.” EIA S/CO ratio is a comparison of the opticaldensityofthepatient’spositiveEIAresulttotheop-ticaldensityofthelaboratory’spositiveEIAcontrol.Ifthe ratioishigh>3.0usingthemostwidelyemployeddiagnos-tickits,ELISA,thepositivepredictivevalue(thatthepatient trulyhasHCVantibodyintheblood)ofthepatient’sresult ishigh,stillemphasizetheimportanceofconfirmingthese resultsusingPCRorRIBA.
Inourresults,wecouldobservethat90%ofthegroup B samples (S/CO ratio > 3) was positive in the PCR test, confirming the serological tests, but four samples did not confirm the results obtained by ELISA; in contrast with groupA,insixtysamplesofgroupB(S/COratio<3)only 8wereconfirmedaspositivebyRT-PCR.Thereasonforthe differenceinresultsbetweenthetechniquesisnotclear.We
cansuggestthatthesepatientshavelowviralloadinwhich wasnotpossibletodetecttheRNA,asourtestshowedthe detectionlimitof310IU/mL.Moreover,therearedatafrom literatureshowingthatsomepatients,whodevelopedanti-HCV,hadclearanceofviralRNAduringacuteinfection.39,40 However,thissubjectneedtobemoreexploredandstudied. Ontheotherhand,ourserviceuseasafetymarginof10% on cut-off, meaning that the samples negative by RT-PCR and positive by ELISA, could be within this margin, con-firmingthedataobtainedbyRT-PCR.
InotherstudiesinBrazil,samplesrepeatedlypositiveto EIA-2 with S/CO ratio > 3 were associated with 100% of true-positiveresultsandhavearound92%ofpositivityfor HCVRNAbyRT-PCR.41
Hence, through our study, we conclude that the One-Step methodology was easy to apply, adding agility to the PCR methods with reliable results, and we can state that thereisastrongpositiverelationshipbetweenS/COandthe resultspresentedbyreal-timePCRinHCVdetection.
DISCLOSURE
All the authors declared no competing interests, and the work content represents the views of co-authors, and that neithertheauthornorthemainco-authorssubmittedman-uscripts duplicate or overlapping in another publication, andthatthearticlementionedinthetextaspersonalcom-municationsareapprovedbythepersonreferenced.
REFERENCES
1. AlterMJTransmissionofHCV–Route,DoseandTiter.New EnglandJournalofMedicine1994;173:784-86.
2. LokASF,GunaratnamNT.DiagnosisofHepatitisC.Hepatol-ogy1997;263:485-565.
3. KrugLP,LungeVR,IkutaN.GenótiposdovírusdahepatiteC (VHC)noRioGrandedoSul.Laes&Haes1997;19:78-82. 4. Justa MTR, Terada C. Experiência do Laboratório Fleury na
elucidaçãodeperfisinconclusivosnasorologiaparaovírusda hepatiteC(HCV).CongressodePatologiaeMedicinaLabora-torial–SãoPaulo,1999.
5. SreevatsanS,BookoutJB,RingpsisFM.AlgorithmicApproach toHigh-throughputMolecularScreeningforAlphainterferon-resistantGenotypesinHepatitisCPatients.JournalofClinical Microbiology1998;36:1895-901.
6. WorldHealthOrganization.Epidemicandpandemicalertand response.Availableat:http://www.who.int/csr/disease/hepati-tis/whocdscssrlyo2003/en/index2.html.AccessedFebruary09, 2009.
7. MchutchinsonJG,GordonSC,SchiffER.InterferonAlpha-2b AloneorinCombinationwithRibavirinasInitialTreatment for Chronic Hepatitis C. Hepatitis Interventional Therapy Group.NewEnglandJournalofMedicine1998;339:1485. 8. GretchDR.UseandInterpretationofHCVDiagnosticTestsin
theClinicalSetting.ClinicsinLiverDisease1997;1:553-7. 9. Muerhoff AS, Jiang L, Shah DOet al. Detection of HCV
10. MedinaM,SchiffER.HepatitisC:DiagnosticAssays.Seminars inLiverDisease1995;15:33-40.
11. LauJYN,DavisGL,KniffenJ,QianK.SignificanceofSerum HepatitisCvirusRNALevelsinChronicHepatitisC.Lancet 1993;341:1501-4.
12. Huber KR, Knapp M, Bauer K. Subtyping Hepatitis CVirus ClinicalChemistry1995;41:319-20.
13. LefrèreJJ,CosteJ,DeferC.ScreeningBloodDonstionforViral Genomes:MulticenterStudyofReal-TimeStimulationUsing PooledSamplesontheModelofHepatitisCVirusRNADe-tection.Transfusion1998;38:915-23.
14. KrugLP,LungeVR,IkutaN.HepatitisCVirusGenotypesin Southern Brazil. Brazilian Journal of Medical and Biological Research1996;29:1629-32.
15. Ohto H, Terezawa S, Sasaki N. Transmission of Hepatitis C VirusfromMotherstoInfants.NewEnglandJournalofMedi-cine1994;17:744-50.
16. PogamSL,DuboisF,ChristenR,RabyC.ComparisonofDNA EnzymeImmunoassayandLineProbeAssays(InnoLipaHCV IandII)forhepatitisCvirusgenotyping.JournalofClinical Microbiology1998;36:1461-3.
17. WangT,LeeSS.HepatitisC:Areviewforprimarycarephysi-cals.CMAJ2006;174:649-59.
18. KamalSM.AcutehepatitisC:asystematicreview.Am.J.Gas-trol2008;103:1283-97.
19. Chevaliez CS, Pawlotsky JM. Practical use of hepatitis C vi-ruskineticsmonitoringinthetreatmentofchronichepatitis. JournalofViralHepatitis2007;14:77-81.
20. FregeauCJ,LettCM,ElliottJ,YensenC,FourneyRM.Auto-matedprocessingofforensiccaseworksamplesusingrobotic workstations equipped with nondisposable tips: contamina-tionprevention.J.Forensic.Sci2008;53:632-51.
21. GerberdingJL,FlemingDW,SniderDE.Guidelinesforlabo-ratorytestingandresultreportingofantibodytohepatitisC virus.EpidemiologyProgramOffice,CentersforDiseasecon-trol and Prevention (CDC), Department of Health and Hu-manServices2003;52:1-15.
22. WhalleySA,MurrayJM,BrownDet al .Kineticsofacutehepa-titisBvirusinfectioninhumans.J.Exp.Med2001;193:847-54. 23. HumarA,KumarD,BoivinB,CaliendoAM.Cytomegalovirus
(CMV)virusloadkineticstopredictrecurrentdiseaseinsol-id-organtransplantpatientswithCMVdisease.J.Infect.Dis 2002;186:829-833.
24. SnijdersPJ,VanDenBrulEAJ,MeijerCJ.Theclinicalrelevance of human papillomavirus testing: relationship between ana-lyticalandclinicalsensitivity.J.Pathol2003;201:1-6.
25. BiedermannK,DandachiN,TrattnerMet al.Comparisonof
real-timePCRsignal-amplifiedinsituhybridizationandcon- ventionalPCRfordetectionandquantificationofhumanpap-illomavirusinarchivalcervicalcancertissue.J.Clin.Microbiol 2004;42:3758-65.
26. WagnerHJ,ChengYC,HulsMHetal.Promptversus preemp-tiveinterventionforEBVlymphoproliferativedisease.Blood 2004;103:3979-81.
27. WatzingerF,SudaM,PreunerSet al.Real–timequantitative PCR assay for detection and monitoring of pathogenic
hu-manvirusesinimmunosuppressedpediatricpatients.J.Clin. Microbiol2004;42:5189-98.
28. Selvin PR. Fluorescence resonance energy transfer. Methods Enzymol1995;246:300-34.
29. HollandPM,AbramsonRD,WatsonR,GelfandDH.Detec- tionofspecificpolymerasechainreactionproductbyutiliz-ingthe50-30exonucleaseactivityofthermosaquaticusDNA polymerase.Proc.Natl.Acad.Sci.USA1991;88:7276-80. 30. BeldM,SentjensR,RebersS.PerformanceoftheNewBayer
VERSANTHCVRNA3.0assayforquantificationofhepati- tisCvirusRNAinplasmaandserum:Conversiontointerna- tionalunitsandcomparisonwiththeRocheCOBASAmpli-corHCVMonitor,version2.0,assay.J.Clin.Microbiol2002; 40:788-93.
31. LeeSC,AntonyA,LeeNet al.Improvedversion2.0qualitative andquantitativeAMPLICORreversetranscription-PCRtests forhepatitisCvirusRNA:Calibrationtointernationalunits, enhancegenotypereactivity,andperformancecharacteristics. J.Clin.Microbiol2000;38:4171-79.
32. KawaiS,YokosubaO,KandaTJ.QuantificationofhepatitisC virusbyTaqManPCR:comparisonwithHCVAmplicorMon-itorassay.J.Med.Virol1999;58:121-6.
33. Komurian-PradelF,Paranhos-BaccalaG,SosoyerM.Quanti-ficationofHCVRNAusingreal-timePCRandfluorimetry.J. Virol.Methods2001;95:111-9.
34. PuigM,MihalikK,YuMY.,FeinstoneSM,MajorME.Sensitive andreproducibilityofHCVquantitationinchimpanzeesera using TaqMan real-tiem PCR assays. J.Virol. Methods 2002; 105:253-63.
35. Takeuchi T, Katsume A, Tanaka Tet al. Real-time detection systemforquantificationofhepatitisCvirusgenome.Gastro-enterology1999;116:636-42.
36. TraniLD,BediniB,DonatelliIet al .Asensitiveone-stepreal-timePCRfordetectionofavianinfluenzavirusesusingaMGB probeandaninternalpositivecontrol.BMC.InfectiousDis-eases2006;87:1-8.
37. ArrudaBC,LiraRA,LoureiroPet al.Evaluationofrealtime PCRtechniquetodiagnosisofhumanT-lymphotropicvirus type I (HTLV-I) in patients in hematology at Fundação He-mope Hospital, in northeastern Brazil. Rev. Bras. Hematol. Hemoter2008;30:384-9.
38. WangLN,WuJM,DengWet al.Lyophilizedstandardsforthe calibrationofrealtimePCRassayforhepatitisCvirusRNA.J. Chin.Med.2006;119:1910-4.
39. MeyerMF,LehmannM,CornbergMet al.Clearanceoflow
levels of HCV viremia in the absence of a strong adaptative immuneresponse.VirologyJournal2007;11:1-11.
40. KondiliLA,ConstantinoA,VillanoUet al.Infectionrateand spontaneous seroreversion of anti-hepatitis C virus during thenaturalcourseofhepatitisCvirusinfectioninthegeneral population.LiverDisease2002;50:693-6.