A Low Stringency Polymerase Chain Reaction Approach to
the Identification of
Biomphalaria
glabrata
and
B.
tenagophila,
Intermediate Snail Hosts of
Schistosoma
mansoni
in Brazil
Teofânia HDA Vidigal/
+, Emmanuel Dias Neto */**, Andrew JG Simpson**
*,
Omar S Carvalho
Laboratório de Helmintoses Intestinais *Laboratório de Parasitologia Celular e Molecular, Centro de Pesquisas René Rachou-FIOCRUZ, Caixa Postal 1743, 30190-002 Belo Horizonte, MG, Brasil **Departamento de Bioquímica e Imunologia, ICB, UFMG, Belo Horizonte, MG, Brasil ***Instituto Ludwig de Pesquisas do
Câncer, Laboratório de Genética do Câncer, São Paulo, SP, Brasil
The low stringency-polymerase chain reaction (LS-PCR) with a pair of specific primers for the am-plification of the 18S rRNA gene was evaluated as a means of differentiating between the two
Schisto-soma mansoni intermediate host species in Brazil: Biomphalaria glabrata and B. tenagophila. Individual snails obtained fromdifferent states of Brazil were used and the amplification patterns obtained showed a high degree of genetic variability in these species. Nevertheless, 4 and 3 clearly defined specific diagnostic bands was observed in individuals from B. glabrata and B. tenagophila respectively. The detection of snail specific diagnostic bands suggests the possibility of reliable species differentiation at the DNA level using LS-PCR.
Key words: polymerase chain reaction - low stringency PCR - Biomphalaria - identification - Brazil
Biomphalaria glabrata and B. tenagophila are
important intermediate snail hosts of Schistosoma mansoni in Brazil although B. straminea also acts
as a host in same areas and B. amazonica and B. peregrina can be infected under artificial
condi-tions (Corrêa & Paraense 1971, Paraense 1973). These snails exhibit extensive intraspecific variation at the morphological (Paraense & Deslandes 1955, Paraense 1975) and genetic level (Knight et al. 1991, Vidigal et al. 1994). Intraspe-cific morphological heterogeneity, in particular, complicates snail identification especially of small specimens. An efficient and reliable method of identifying species, would be extremely valuable in the study of the distribution host species both in areas of active transmission of schistosomiasis and non endemic areas, were the presence of suscep-tible species should be notified to alert the health services. Because of its simplicity and sensitivity, a snail identification test based on the polymerase chain reaction (PCR) would be ideal. As a step in
This study was supported in part by CNPq and FAPEMIG.
+Corresponding author. Fax: +55-31-295.3115 Received 26 January 1996
Accepted 15 May 1996
this direction two groups (Langand et al. 1993, Vidigal et al. 1994) have independently undertaken the amplification of host snail DNA using the ar-bitrarily primed PCR (AP-PCR) (Welsh & McClelland 1990), in order to produce randomly amplified polymorphic DNAs, RAPDs (Williams et al. 1990). However, the results obtained by Vidigal et al. (1994) working within B. glabrata,
showed a remarkable degree of intraspecific poly-morphism, with the result that the RAPD patterns observed were so variable, that there were no bands common to all the specimens analyzed. This sug-gested that an AP-PCR based test for species iden-tification would be extremely difficult to develop. As an alternative to AP-PCR we have here ex-plored the related methodology of low stringency-PCR (LS-stringency-PCR) where instead of arbitrarily selected primers, primers that specifically amplify defined regions of the genome, are used under the amplifi-cation conditions used for AP-PCR (Dias Neto et al. 1993). The result of LS-PCR amplification is a specific fragment defined by the primer used, to-gether with a complex set of other fragments (known as low stringency products or LSPs) de-rived from low stringency interactions of the prim-ers with other sequences in the target genome. This method has been of value in S. mansoni sex
quantifi-cation of human papilomavirus infection (Cabal-lero et al. 1995). In the present work we elected to use rRNA gene specific primers for LS-PCR with the idea that since the rRNA gene is highly con-served, related sequences amplified by LS-PCR may also exibit a greater intraspecific stability than randomly amplified sequences. The results ob-tained were consistent with this hypothesis in that a number of B. glabrata and B. tenagophila
spe-cific amplification products were identifiable from all specimens of these two species studied provid-ing the basis of a PCR identification test. The meth-odology thus offers promise as a molecular ap-proach to snail identification in the context of the epidemiology and control of schistosomiasis mansoni.
MATERIALS AND METHODS
Snail populations - The snail specimens
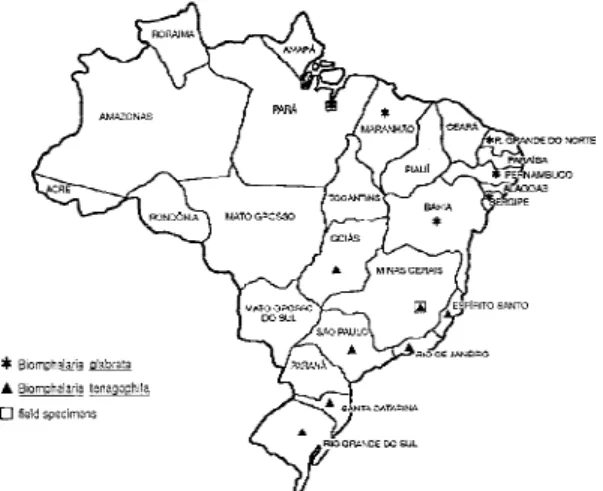
stud-ied were maintained at the Department of Mala-cology of the Institute Oswaldo Cruz, Rio de Janeiro, with the exception of the specimens of B. glabrata from Belém, PA andB. tenagophila from
Vespasino, MG, which were collected directly from the field. The studies were undertaken using populations obtained from localities shown in the Fig. 1. The dates and the sites of collection as well as the number of specimens in the original samples are shown in Table. All the snails used in the study were reared and maintained at room temperature under identical conditions in aquarium with run-ning water and calcium carbonate as substrate. The snails were exposed to artificial diurnal lighting of 10 hr and fed with lettuce. In all cases, the snails were identified by means of comparative morphol-ogy based on the reproductive organs and shells (Paraense l975) and examined for the presence of infection by S. mansoni. None of the snails were
found to be infected.
DNA extraction - Total DNA was extracted
from the foot of the snails basically as described previously (Vidigal et al. 1994). Briefly, the foot of each snail was mechanically disrupted in 50mM Tris HCl pH 8.0, 100 mM NaCl, 50 mM EDTA, 0.5% SDS and incubated with 50µg/ml proteinase K overnight at 37ºC. Following phenol/chloroform extraction and ethanol precipitation, DNA was re-suspended in 10 mM Tris-HCl, 1 mM EDTA pH 8.0 and the DNA concentration estimated by com-parison with known standards on 2% ethidium bro-mide stained agarose gels.
The LS-PCR technique - The primers used were
based on the sequence of the 18s rRNA gene from the bivalve mollusk Placopecten available in
GenBank (access number X53899). The primers correspond to a conserved region of the gene and show a high degree of homology with human (ac-cess number X03205), S. mansoni (access number
X53467), fungi (Neurospora crassa, access
num-ber X04971) and other 18s rRNA genes.
The protocol used was that previously applied to the sexual determination of human DNA and larval stages of S. mansoni (Dias Neto et al. 1993).
DNA samples from each individual were ampli-fied in duplicate using 100 pg and 1 ng of template DNA. Each reaction was undertaken in final vol-ume of l0µl containing 0.8 units of Taq DNA poly-merase (Cenbiot RS, Brazil), 200 µM of each dNTP, 1.5 mM MgCl2, 50mM KCl, 10mM Tris-HCl pH 8.5, together with 6.4 pmoles of the primers NS1 (5' GTAGTCATATGCTTGTCTCAG -3') and ET1 (5'- GTCCAGACACTACGGGAAT - 3'). The reaction mix was overlaid with 20µl of mineral oil and, following an initial denaturation at 95°C for 5 min, was subjected to two cycles through the following temperature profile: 30°C for 2 min for annealing, 72°C for 1 min for exten-sion and 30 sec at 95°C for denaturation followed by 33 cycles where the annealing step was altered to 40°C. In the final cycle, the extension step was for 5 min. Following amplification, 3µl of each reaction was mixed with 1µl of 4x sample buffer (0.125% bromophenol blue, 0.125% xylene cyanol, 15% glycerol) and subjected to electrophoresis in 4% polyacrylamide gel (acrylamide/bisacrylamide 29:1) in TBE buffer (2 mM EDTA,10 mM Tris borate pH 8.0). The gels were silver stained by fix-ing with 10% ethanol/ 0.5% acetic acid for 3 min, staining with 0.2% silver nitrate in the fixing solu-tion for 5 min and reducsolu-tion with 0.75 M NaOH/ 0.1M formaldehyde for 5 min (Sanguinetti et al. 1994).
High stringency amplification - For specific
amplification of the 18s rRNA region, 3 pmol of each primer (NS1 and ET1) and 0.4 units of Taq DNA polymerase (Cenbiot RS, Brazil) per l0µl
reaction mixture were used. The other components of the reaction mixture were as described above for LS-PCR. The specific amplification consisted of an initial cycle with denaturation step at 95°C for 5 min, annealing of primers at 60°C for 2 min and 72°C for 2 min for extension, this was fol-lowed by 29 cycles of amplification at 60°C for 2 min, 72°C for 2 min, and 95°C for 45 sec, with an extended incubation at 72°C for 5 min in the final cycle.
RESULTS
Specimens of B. glabrata and B. tenagophila,
derived from various geographical regions were analyzed (Table). In total six populations of B. glabrata and seven of B. tenagophila were
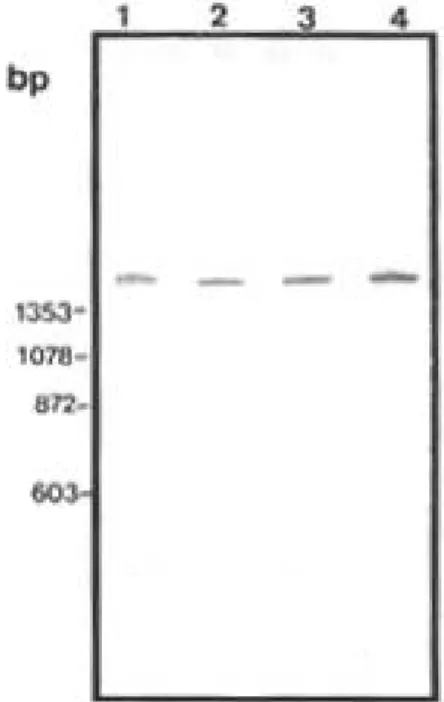
stud-ied.When the primer pair NS1 and ET1 used un-der high stringency amplification conditions, only the specific band, of approximately 1500 bp cor-responding to the 18s rRNA gene, was seen in all individuals of these species, indicating the absence of detectable size polymorphism in this gene in the organismsstudied (Fig. 2).
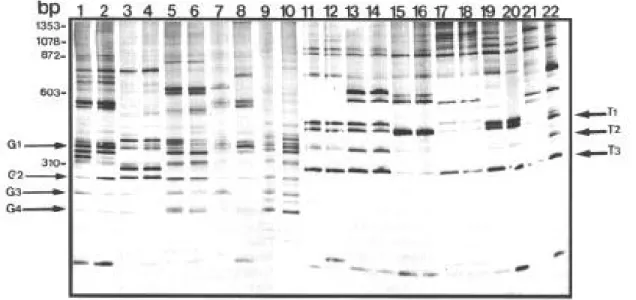
When LS-PCR was used, complex patterns were produced composed of low stringency prod-ucts (LSPs) derived from multiple interactions of the primer pair throughout the genomes studied (Fig. 3). Due to the competitive nature of LS-PCR (Caballero et al. 1995) the specific bands shown in Fig. 2 were of weak and variable intensity. The pattern of LSPs was polymorphic for both species however, a number of bands were consistently amplified from all the specimens of each species. The result shown in Fig. 3 is an experiment that formed part of a screening procedure aimed at iden-tifying primers capable of distinguishing B. glabrata and B. tenagophila. The B. glabrata
specimens were characterized by LS-PCR with the
primers NS1/ ET1 by the presence of one or two LSP doublets between 370 and 480 bp (G1) and strong LSPs of 290, 260 and 240 bp (G2, G3 and G4). The B. tenagophila specimens were
charac-terized by a clear LSP doublet of 500 bp (T1), strong LSPs of 400 and 310 bp (T2 and T3) as well as the absence of major LSPs between 290
TABLE
Places and dates of Biomphalaria populations
Species Origin Site Date of collection
B. glabrata Belém, PA Stream November, 1992
B. glabrata Cururupu, MA River May, 1989
B. glabrata Touros, RN Lake November, 1975
B. glabrata Pontezinha, PE Stream April, 1989
B. glabrata Aracaju, SE Stream November, 1985
B. glabrata Jacobina, BA River June, 1984
B. tenagophila Paracambi, RJ Stream May, 1990
B. tenagophila Imbé, RS NA NA
B. tenagophila Araçatuba, SP NA May,1981
B. tenagophila Formosa, GO NA August, 1981
B. tenagophila Vespasiano, MG Stream May, 1994
B. tenagophila Joinvile, SC NA NA
B. tenagophila Vila Velha, ES NA January, 1983
NA: data not available
and 240 bp.
In order to test the generality of specific bands for the two species the number of specimens was increased (Fig. 4) The major banding characteris-tics described in Fig. 3 were generally maintained for both species. The pair of LSP doublets (G1) appeared as a variable complex of bands. Although the precise details of the complex were variable it was present in one form or another in all B. glabrata
specimens and absent from all B. tenagophila
speci-mens. The other B. glabrata bands were
consis-tent in all specimens (although with variable in-tensity) and absent from all the B. tenagophila
specimens. For B. tenagophila the most useful
char-acters were T3 and the absence of detectable LSPs between 290 and 240 bp. T2 was present in all specimens at variable intensity but two of the B. glabrata specimens (lanes 5 and 6) also exhibited
a strong LSP of this size. The doublet T1 was not consistent although at least one band of the same size was present in all the specimens tested and in none of the B. glabrata specimens were
corre-sponding LSPs detectable.
On the basis of these results, a double blind test was undertaken using of eight specimens col-lected directly from the field, in the town of Vespasiano, MG, as a test of the LS-PCR identifi-cation system (Fig. 5). Individual 1 made the
mor-Fig. 3: silver-stained 4% polyacrylamide gel showing the LS-PCR products obtained with primers NS1 - ET1 and 1ng of DNA from snails. Lane 1: Biomphalaria. glabrata from Belém, PA. Lane 2: B. glabratafrom Pontezinha, PE. Lane 3: B. glabrata from Aracaju, SE. Lane 4: B. glabrata from Jacobina, BA. Lane 5: B. tenagophila from Paracambi, RJ. Lane 6:B. tenagophila from Imbé, RS. Lane 7: B. tenagophila from Formosa, GO. Lane 8: B. tenagophilafrom Araçatuba, SP. The species diagnostic LSPs are indicated by arrows, and the molecular weight markers are as shown.
phological characterization and the DNA extrac-tion. The DNA was then amplified and the diag-nostic LSPs evaluated by individual 2. When this profile was compared with the position of the char-acteristic bands for the two species as marked on the figure the samples were identified as B. tenagophila. The most conclusive features were the
presence of T1 and T2, and the absence of bands between 290 and 240 bp. Band T3 is present in all samples although it is considerably weaker in lanes 5 to 7. The DNA based identification was con-firmed by the analysis of morphological charac-ters.
DISCUSSION
The classical methods of identification of fresh-water snails of medical importance is difficult be-cause of the variation encountered in the anatomi-cal and morphologianatomi-cal characteristics commonly used for separating species (Paraense 1975, Jelnes 1979, 1986, Henricksen & Jelnes 1980). The analy-sis of anatomy of the reproductive organs and mor-phological characters of the shell can be time-con-suming when several specimens are to be exam-ined. This traditional morphological characters, used by malacologists, for identification of spe-cies, have not always been easily utilized by non-specialist. Thus, an easy and reliable method for species identification is important in efforts to con-trol schistosomiasis.
Techniques of molecular biology have recently been used to study the variability of basom-matophora mollusks and to identify species of
Bulinus and Biomphalaria snails. This
methodol-ogy allows the direct and detailed examination of the genome and opens up new possibilities for snail identification and phylogenetic studies (Knight et al. 1991, Rollinson & Kane 1991, Strahan et al. 1991, Langand et al. 1993, Vidigal et al. 1994).
Langand et al. (1993) using RAPD analysis demonstrated the potential of this technique for differentiating populations and species of Bulinus.
Vidigal et al. (1994) showed that the genome of B. glabrata exhibits a remarkable degree of
intraespecific polymorphism. All the bands that were amplified by two randomly chosen primers were found to be polymorphic. Those results sug-gested that RAPD analysis would not be appropri-ate for identifying relappropri-ated species of Biomphalaria.
The present study revealed however that LS-PCR using primers for 18s rRNA does allow the identification of B. glabrata and B. tenagophila.
The study of individuals obtained from geographi-cally distinct populations (Fig. 1) demonstrated in-traspecific DNA polymorphism by LS-PCR, how-ever specimens from the same species, exhibited some species specific LSPs. The fact that the popu-lations used for this study were obtained from widely separated localities indicate that these prim-ers should allow the identification of B. glabrata
and B. tenagophilathroughout Brazil.
Preliminary experiments with populations of B. straminea using the same methodology suggests
that this species is even more variable than B. glabrataandB. tenagophila, no bands are shared
between all specimens studied. However at a re-gional level, shared LSPs were found permitting specific identification in this case (data not shown). Most importantly, B. straminea DNA did not, when
used as a template for LS-PCR with the 18s rRNA primers, generate any of the diagnostic LSPs used for identification of B. glabrata and B. tenagophila.
Thus, although the methodology cannot be used to conclusively identify B. straminea this snail will
not be incorrectly identified as either B. glabrata
or B. tenagophila.
In summary, the data show that LS-PCR analy-sis is appropriate for distinguishing B. glabrata
from B. tenagophila. The technique requires small
amounts of DNA and can be applied to juvenile snails as a means of resolving problems of identi-fication of potential intermediate snail hosts in Brazil, in situations where the classical taxonomy methodology is inconclusive.
ACKNOWLEDGMENTS
To Dr Wladimir Lobato Paraense and Dr Ligia Corrêa from the Departamento de Malacologia, Fundação Oswaldo Cruz, for giving us snails and Dr Izabel de Carvalho Rodrigues from the Instituto Evandro Fig. 5: silver-stained 4% polyacrylamide gel showing the
Chagas, Fundação Nacional de Saúde, for giving us snails from Belém, Pará. To Dr Álvaro Romanha for making available the facilities used in the development of the project and José Geraldo Amorim da Silva and Maria Ângela A dos Santos for their technical assistance.
REFERENCES
Caballero OLSD, Dias Neto E, Koury MC, Romanha AJ, Simpson AJG 1994. Low-Stringency PCR with diagnostically useful primers for identification of
Leptospira serovars. J Clin Microbiol32:
1369-1372.
Caballero OLSD, Villa LL, Simpson AJG 1995. Low stringency PCR (LS-PCR) allows entirely internally standardized DNA quantitation. Nucleic Acids Res 23: 362-363.
Corrêa LR, Paraense WL 1971. Susceptibility of
Biomphalaria amazonica to infection with two
Bra-zilian strains of Schistosoma mansoni. Rev Inst Med Trop São Paulo 13: 387-390.
Dias Neto E, Santos FR, Pena SDJ, Simpson AJG 1993. Sex determination by Low Stringency PCR (LS-PCR). Nucleic Acids Res 21: 763-764.
Henricksen UB, Jelnes EJ 1980. Experimental taxonomy of Biomphalaria (Gastropoda: Planorbidae). I. Meth-ods for experimental taxonomic studies on
Biomphalaria carried out by horizontal starch gel
electrophoresis and staining of twelve enzymes. J Chromatogr 188: 169-176.
Jelnes EJ 1979. Experimental taxonomy of Bulinus (Gas-tropoda: Planorbidae). II. Recipes for horizontal starch gel electrophoresis of ten enzymes in Bulinus
and description of internal standard systems and two new species of the Bulinus forskalii complex. J Chromatogr170: 405-411.
Jelnes EJ 1986. Experimental taxonomy of Bulinus (Gas-tropoda: Planorbidae): the West and North African species reconsidered, based upon an electrophoretic study of several enzymes per individual. Zool J Linn Soc87: 1-26.
Knight M, Brindley PJ, Richards CS, Lewis FA 1991.
Schistosoma mansoni: Use of a cloned ribosomal
RNA gene probe to detect restriction fragment length polymorphisms in the intermediate host
Biomphalaria glabrata. Exp Parasitol 73:285-294.
Langand J, Barral V, Delay B, Jourdane J 1993. Detec-tion of genetic diversity within snail intermediate hosts of the genus Bulinus by using random
ampli-fied polymorphic DNA markers (RAPDs). Acta Trop 55: 205-215.
Paraense WL 1973. Susceptibility of Biomphalaria peregrina from Brazil and Ecuador to two strains
of Schistosoma mansoni. Rev Inst Med Trop São Paulo 15: 127-130.
Paraense WL 1975. Estado atual da sistemática dos planorbídeos brasileiros. Arq Mus Nac Rio de Janeiro 55:105-128.
Paraense WL, Deslandes N 1955. Observations on the morphology of Australorbis glabratus. Mem Inst Oswaldo Cruz 53: 87-103.
Rollinson D, Kane RA 1991. Restriction enzyme analy-sis of DNA from species of Bulinus
(Basom-matophora; Planorbidae) using a cloned ribosomal RNA gene probe. J Moll Stud 57: 93-98.
Sanguinetti CJ, Dias Neto E, Simpson AJG 1994. Rapid silver staining and recovery of PCR products sepa-rated on polyacrylamide gels. BioTechiniques 17: 915-918.
Strahan K, Kane RA, Rollinson D 1991. Development of cloned DNA probes for the identification of snail intermediate hosts within the genus Bulinus. Acta Trop 48: 117-126.
Vidigal THDA, Dias Neto E, Carvalho OS, Simpson AJG 1994. Biomphalaria glabrata: extensive genetic
varia-tion in Brazilian isolates by random amplified poly-morphic DNA analysis. Exp Parasitol 79: 187-194.
Welsh J, McCleelland M 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18: 7213-7218.
Williams JGK, Kubelick AR, Livak KJ, Rafalski JA, Tingey SV 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers.