Genetic diversity and phylogenetic relationships in the rye genus
Secale
L.
(rye) based on
Secale cereale
microsatellite markers
Hai-Ying Shang
1*, Yu-Ming Wei
1,2*, Xiao-Rong Wang
1, You-Liang Zheng
1,21
Triticeae Research Institute, Sichuan Agricultural University, Yaan, Sichuan, China.
2
Key Laboratory of Crop Genetic Resources and Improvement in Southwest China, Ministry of Education,
Sichuan Agricultural University, Yaan, Sichuan, China.
Abstract
The genetic diversity and phylogenetic relationships in the genusSecale L. (rye) was evaluated using 24 Secale cereale microsatellite (SCM) markers. The average polymorphism information content (PIC) value of each microsatellite locus in 30Secale accessions evaluated was higher than that in 47 cultivated ryes (Secale cereale ssp. cereale). The mean genetic similarity (GS) index in Secale was lower than that in cultivated rye. The highest within-species GS index was observed forS. sylvestre and the lowest for S. strictum, whereas the highest be-tween-species GS index was found betweenS. cereale and S. vavilovii and the lowest between S. sylvestre and S. cereale. There was no obvious difference in GS levels in the cultivated rye accessions from Asia, Europe, North America or South America. Cluster analysis indicated that all theSecale accessions could be distinguished by the 24 microsatellite loci. We also found that theS. sylvestre accessions were obviously divergent from the accessions of other species and that theS. vavilovii accessions were closely related to the S. cereale accessions. Our results also showed thatS. strictum was heterogeneous and showed great within-species differences. The microsatellite-derived dendrogram faithfully reflected the phylogenetic relationships betweenSecale species but did not indicate a possible domestication process of the cultivated rye based on the geographical sources of the accessions.
Key words:cultivated rye, genetic diversity, microsatellite markers, phylogenetic relationships,SecaleL. Received: August 23, 2005; Accepted: March 15, 2006.
Introduction
The taxonomy of the genusSecale(rye) has for a long time been a matter of disagreement, despite the large num-ber of studies performed. Historically, about 15 different species have been accepted (Roshevitz 1947, Delipavlov 1962), while Frederiksen and Petersen (1998) recognized only threeSecalespecies,i.e. S. sylvestre,S. strictum(plus the subspecies ssp.strictumand ssp.africanumand the va-rieties var.strictumand var.ciliatoglume) andS. cereale (plus ssp.cerealeand ssp.ancestrale). According to taxo-nomic systems adopted by the American Germplasm Re-sources Information Network (GRIN, http://www. ars-grin.gov), the genusSecaleis presently recognized as containing four species, consisting of the annual outbreeder S. cerealeL. , the annual autogamousS. sylvestreHost and S. vavilovii Grossh., and the perennial outbreeder S. strictum (Presl.) Presl. (syn. S. montanum) (Sencers and Hawkes 1980, De Bustos and Jouve 2002). There are 8
sub-species inS. cerealeand 5 inS. strictum, andS. cerealessp. cerealeis the only cultivated rye.
The different recent classifications of the genus Secalehave involved phylogenetic analyses based on the use of various technologies, including some biochemical and molecular markers such as isozymes (Vences et al. 1987a,b), random amplified polymorphic (RAPDs) (Del Pozoet al. 1995), rDNA spacer-length (Reddyet al. 1990, Cuadrado and Jouve 2002) and internal transcribed spacers (ITS) (De Bustos and Jouve 2002) as well as traditional morphological and cytogenetical methods (De Bustos and Jouve 2002). Simple sequence repeats (SSRs or micro-satellites) consisting of tandem repeats of 2 to 6 nucleotides are abundantly distributed throughout the nuclear genomes of all studied plant species (Tautzet al. 1986, 1989, Litt and Luty 1989). Because of their co-dominant inheritance, high polymorphism, good reproducibility and the convenience of the polymerase chain reaction (PCR) microsatellites have become the genetic markers of choice for studies in-volving plant species (Powellet al. 1996, Zhebentyayevaet al. 2003). Takezake and Nei (1996) concluded that micro-satellite DNA seemed to be very useful for clarifying the
www.sbg.org.br
Send correspondence to You-Liang Zheng. Triticeae Research In-stitute, Sichuan Agricultural University, Yaan, Sichuan 625014, China. E-mail: ylzheng@sicau.edu.cn. *These authors contributed equally to this paper.
evolutionary relationships of closely related populations. However, the application of microsatellite markers to plant taxonomy is still very limited because the development of specific SSR primers is time-consuming and expensive plus the fact that genomic microsatellite markers are hard to transfer between the genomes of different species. Fortu-nately, at least 184S. cerealemicrosatellite (SCM) markers have recently been developed (Saal and Wricke 1999, Ha-ckauf and Wehling 2002).
The objectives of the study described in the present paper were to investigate the phylogenetic relationship of four Secale species using microsatellite markers and to evaluate the genetic diversity and genetic relationships of cultivated rye (S. cerealessp.cereale) from different coun-tries.
Materials and Methods
Plant materials
For this study we selected 30Secaleaccessions, con-sisting of 5 cultivated rye and 25 non-cultivated rye, to rep-resent the 4 species and 10 subspecies of the rye genus (Table 1). Meantime we used 47 cultivated rye (S. cereale ssp. cereale) accessions from different countries (Table 1). Accession CN31389 was kindly provided by Dr. D. Kessler of Plant Gene Resources of Canada (PGRC). Accession NGB5073 was kindly provided by Dr. L. Bondo at Nordic Gene Bank, Sweden. Accessions R953/90 and R955/90 were kindly provided by Dr. A. Graner at the Genebank Gatersleben, Institute of Plant Genetic and Crop Plant Re-search (IPK). Accessions As3045 and As3033 came from the Triticeae Research Institute, Sichuan Agriculture Uni-versity (TRISAU). Most of the accessions with accession numbers starting with PI were kindly provided by Dr. H. Bockelman of the American Germplasm Resources Infor-mation Network (GRIN).
DNA isolation and PCR amplification
Genomic DNA was extracted from young leaves us-ing the cetyltrimethylammonium bromide (CTAB) proce-dure of Doyle and Doyle (1987). The samples for each accession consisted of DNA bulked from 25-30 individual plants.
We used 24 microsatellite markers (Table 2), consist-ing of 13 markers described by Saal and Wricke (1999) and 11 markers described by Hackauf and Wehling (2002). All the primers were synthesized by Shenergy Biocolor, Bio-logical Science and Technology Co., China. The final reac-tion volume was 25 mL, containing approximately 50-100 ng template DNA, 1 unit of Takara rTaq polymer-ase (Takara Bio, Inc., Kyoto, Japan), 0.2mM of each pri-mer, 200 mM of each deoxynucleotide triphosphates (dNTP) (Takara Bio, Inc., Japan), 1.5 mM MgCl2, and
1xPCR buffer. The PCR amplifications were carried out in a PTC-240 thermocycler (Genetic Technologies, MJ
Re-search, USA) under the conditions described by Saal and Wricke (1999) and Hackauf and Wehling (2002) with mi-nor modification. The PCR amplification products were separated on a 6% (w/v) denatured polyacrylamide gel and visualized by silver staining.
Data scoring and analysis
Polymorphism information content (PIC) values were calculated for each microsatellite locus according to the formula:
PICi Pij j
n =
-=
å
1 2
1
wherepijis the frequency of thejth allele for theith marker
summed overnalleles (Andersonet al. 1993). For each ge-notype x marker combination, the presence (1) or absence (0) of a microsatellite allele was treated as an independent character. The data matrix was then used to calculate the genetic similarity (GS) index (Nei and Li 1979) as GS= 2Nij/(Ni+Nj), whereNijis the number of microsatellite
alleles common to genotypesiandj, whileNiandNjare the
total numbers of microsatellite alleles observed for geno-types i and j, respectively. Genetic relationships among Secale accessions were estimated using the unweighted pair-group method with arithmetic mean (UPGMA) cluster analysis of theGSmatrix (Rohlf, 2000)
Results
Five cultivated rye (S. cereale ssp. cereale) acces-sions and 25 accesacces-sions of otherSecalespecies or subspe-cies were selected to detect the genetic variations and rela-tionships within and betweenSecale species. For the 30 accessions selected a total of 113 microsatellite alleles were amplified, of which 107 (94.7%) were polymorphic with an average of 4.7 alleles per locus and a range of from 2 to 8. The average PIC value of the 13 markers from Saal and Wricke (1999) was 0.589 while that of the 11 markers from Hackauf and Wehling (2002) was 0.620. The average PIC value of the 24 microsatellite loci was 0.604 and ranged from 0.315 to 0.799. In these accessions the highest PIC value was for the SCM101 marker. These results indicated a high level of the microsatellite polymorphism in the 30 accessions
be-Table 1- The 72Secaleaccessions used in this study.
Secale accessions (n = 72)
Cultivated rye (n = 47) Non-cultivated rye (n = 25)
Taxon and accession number
Country, State Continent Taxon and
accession number
Country, State Continent
S. cerealessp. cereale S. cerealessp. afghanicum
CIse 20 Sweden, Malmohus Europe PI618662 Armenia Asia
CIse 26 Canada, Saskatchewan North America
CIse 35 United States North America S. cerealessp. ancestrale
CIse 79 Australia Oceania PI618663 Turkey Asia
PI168199** Turkey, Isparta Asia PI618666 Turkey Asia
PI221478 Afghanistan, Bamian Asia
PI240676 Argentina, Pico South America S. cerealessp. dighoricum
PI254806 Austria, Lower Austria Europe PI618667 Sweden Europe
PI260055 Ukraine, Kharkiv Europe PI618668 Russian Federation Europe
PI272333 Hungary, Heves Europe
PI280839 Russian Federation, Kirov Europe S. cerealessp. rigidum
PI289827 Pakistan Asia PI618669 Turkey Asia
PI290420 Hungary, Pest Europe
PI290423 Slovakia, West Slovakia Europe S. cerealessp. segetale
PI306488 Romania Europe PI618671 Turkey Asia
PI323449 Poland, Lublin Europe PI618673 Turkey Asia
PI330422 Poland Europe
PI344983 Yugoslavia, Montenegro Europe S. cerealessp. tetraploidum
PI345001 Macedonia Europe PI573647 Ukraine, Kharkiv Europe
PI346416** Australia Oceania
PI362399 Yugoslavia, Montenegro Europe S. vavilovii
PI372116 Belarus, Minsk Europe PI573649 Afghanistan Asia
PI372117 Ukraine, Chernihiv Europe PI618678 Armenia Asia
PI372118 Russian Federation, Leningrad Europe PI618680 Turkey Asia
PI372119 Belarus Europe
PI410534 Pakistan, Azad Kashmir Asia S. sylvestre
PI410800 Poland Europe PI592294 Ukraine, Mykolayiv Europe
PI412949** South Africa, Cape Africa NGB5073 Denmark Europe
PI414080 United Kingdom, England Europe R953/90 Denmark Europe
PI430003 India, Himachal Pradesh Asia R955/90 Denmark Europe
PI430004 India, Himachal Pradesh Asia CN31389 Germany Europe
PI436188 Chile, Los Lagos South America AS3045
PI436192 Chile, Los Lagos South America
PI446020 Japan Asia S. strictum
PI446025 Mexico North America PI401400 Iran, Kordestan Asia
PI446366 Bulgaria Europe PI531829 Armenia Asia
PI446379 Ukraine, Lviv Europe PI568257 Russian Federation Europe
PI446433 Bulgaria Europe
PI447337** China, Xinjiang Asia S. strictum ssp. africanum
PI452132 China, Guizhou Asia As3033 South Africa, Calvinia Africa
PI534981 Japan Asia
PI535003 Finland Europe S. strictumssp.anatolicum
PI535005 Finland Europe PI445973 United States North America
PI535018 Portugal Europe PI445974 Canada, Manitoba North America
PI542468 Mexico, Sonora North America
PI573634 Armenia, Syunik Asia S. strictumssp. kuprijanovii
PI602997** United States, Georgia North America PI326282 Russian
Federa-tion, Krasnodar
Europe
tweenS. sylvestreandS. vaviloviiand betweenS. sylvestre andS. strictumare also lower. The results indicated that the S. sylvestreaccessions studied were more divergent from the accessions of the other species. TheGSindex between S. vaviloviiandS. cerealewas the highest at 0.783, indicat-ing thatS. vaviloviiwas closely related toS. cereale.
The genetic relationships inSecaleas estimated using UPGMA cluster analysis of the genetic distance 1-GS ma-trix are shown in Figure 1. All 30 accessions could be dis-tinguished by 24 microsatellite markers. The six S. sylvestreaccessions were divergent from the other acces-sions and all closely clustered into group V. TheS. strictum
ssp.africanumaccession As3033 andS. strictumaccession PI401400 were divergent from the otherS. strictum acces-sions and clustered into group IV, whileS. strictum acces-sions (PI531829 and PI568257) were clustered into group III. TheS. strictumssp.kuprijanoviiaccession PI326282 and twoS. strictumssp.anatolicumaccessions (PI445973 and PI445974) were clustered into group II. The threeS. vaviloviiaccessions and allS. cerealesubspecies were ge-netically related and clustered into group I.
For the 47S. cerealessp.cereale(cultivated rye) ac-cessions (Table 2) a total of 82 microsatellite alleles were amplified, of which 69 (84%) were polymorphic with an
Table 2- Repeat motif, number and size range of alleles, polymorphic information content (PIC) value and chromosome location of theSecale microsatellite markers used in this study. The markers came from the studies of Saal and Wricke (1999) and Hackauf and Wehling (2002).
Marker* Repeat motif Observed
alleles
Size range (bp) PIC in cultivated rye
PIC inSecale Chromosome location
Saal and Wricke (1999)
SCM2 (GT)10 6 110-147 0.146 0.507 6RL
SCM5 (GA)16 7 190-331 0.797 0.789 3RL
SCM9 (GT)8 5 190-242 0.506 0.606 1RS
SCM39 (GT)8(GC)6(GT)53 3 190-331 0.427 0.599 1R
SCM43 (GT)11 4 67-147 0.156 0.616 2R
SCM86 (GT)20 3 110-147 0.449 0.638 7R
SCM101 (CT)18 8 147-190 0.647 0.799 6R
SCM120 (AC)10 5 110-147 0.580 0.556 5RL
SCM138 (AC)23 6 110-190 0.434 0.681 5RS
SCM180 (GT)6(GA)7 2 110-190 0.000 0.358 6RL
SCM304 (GA)36 3 190-331 0.000 0.315 6RS
WMS6 (GA)40 4 110-190 0.358 0.700 5RL
WMS44 (GA)28 2 67-147 0.473 0.498 6R
Hackauf and Wehling (2002)
SCM63 (CCG)5 6 110-190 0.521 0.521 Unknown
SCM66 (CCG)7 5 147-190 0.506 0.680 Unknown
SCM92 (GCG)6 4 147-190 0.698 0.633 Unknown
SCM98 (CTG)5 6 110-147 0.688 0.727 Unknown
SCM116 (TAT)7 3 110-190 0.489 0.464 Unknown
SCM126 (AACC)4 5 110-190 0.780 0.744 Unknown
SCM152 (AG)7 8 190-331 0.558 0.558 Unknown
SCM153 (AT)9 6 147-190 0.764 0.779 Unknown
SCM164 (CCT)5 7 67-190 0.496 0.700 Unknown
SCM166 (CCT)5 6 190-331 0.611 0.649 Unknown
SCM169 (CGG)7 4 147-190 0.230 0.368 Unknown
*Grouped by citing author (see reference list).
Table 3- Genetic similarities (GS) and range of variations within and betweenSecalespecies.
Species S. cereale S. vavilovii S. sylvestre S. strictum
S. cereale 0.779 (0.696 to 0.885)
S. vavilovii 0.783 (0.705 to 0.867) 0.812 (0.764 to 0.817)
S. sylvestre 0.444 (0.326 to 0.543) 0.487 (0.419 to 0.554) 0.884 (0.822 to 0.932)
average of 3.3 alleles per locus and a range of from 1 to 7. We also found that 22 out of the 24 (91.67%) microsatellite markers used were polymorphic in the 47 cultivated rye ac-cessions studied, the remaining two microsatellite markers (SCM 180 and SCM304) being monomorphic. The average PIC value of the 13 markers from Saal and Wricke (1999) was 0.383 while that of the 11 markers from Hackauf and Wehling (2002) was 0.576. In cultivated rye the average PIC value of the 24 microsatellite loci was 0.471 and ranged from zero for SCM180 and SCM304 to 0.797 for SCM5. These results indicate that the microsatellite poly-morphism in the cultivated rye was lower.
The mean GS index for the 47 cultivated rye acces-sions was 0.773 and ranged from 0.622 to 0.921. The high-est GS index (0.921) was observed between the Australian accession PI346416 and accession PI372118 from the Rus-sian Federation, while the lowest (0.622) was between ac-cessions PI542468 from Mexico and PI345001 from Macedonia. The genetic similarity and range of variation between cultivated rye accessions from different continents were calculated based on the GS matrix (Table 4). TheGS index for the European accessions was the highest (0.794)
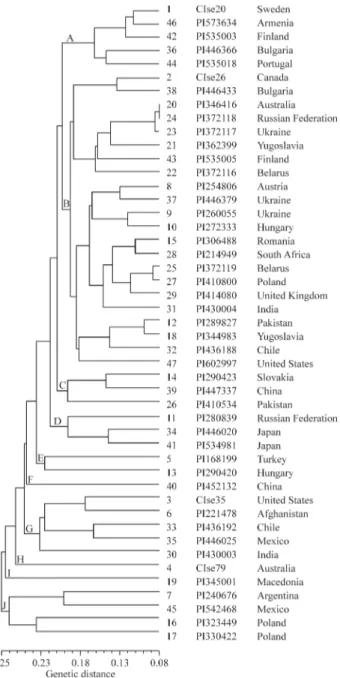
while that for the North and South American accessions was the lowest (0.731). The highestGSvalue (0.777) was observed between Asia and Europe, and the lowest (0.742) between Europe and North American. Our micro-satellite-derived data showed no obvious differences inGS index between the cultivated ryes from different continents. The genetic relationships in cultivated rye (Figure 2) were estimated by UPGMA cluster analysis of the genetic distance (1-GS) matrix. Accessions PI323449 and PI330422 from Poland, PI542468 from Mexico and PI240676 from Argentina clustered in group J and were the most divergent from other accessions, although accessions PI345001 from Macedonia, CIse79 from Australia and PI452132 from China each formed a separate cluster (groups I, H and F respectively) and were also divergent from the other groups. Accessions PI430003 from India, PI221478 from Afganistan, PI446025 from Mexico, CIse35 from United States, and PI436192 from Chile were genetically related and clustered into group G, while acces-sion PI168199 from Turkey and PI290420 from Hungary formed group E. Accessions PI446020 and PI534981 from Japan were related to accession PI280839 from the Russian Federation and clustered into group D, while accession PI447337 from China, PI290423 from Slovakia and PI410534 from Pakistan formed group C. Accessions CIse20, PI446366, PI535003 and PI535018 from Europe and accession PI573634 from Asia clustered into group A. And all the remaining accessions were clustered into one group B.
Discussion
The average microsatellite marker PIC index was 0.604 for the 30Secaleaccessions of different species or subspecies, whereas for the cultivated rye accessions the average PIC index was 0.471. The meanGSindex for the 30Secaleaccessions of different species or subspecies was 0.633 while for the 47 cultivated rye accessions the mean GSindex was 0.773. These results suggest that the genetic diversity in theSecaleas a whole is more extensive than that in cultivated ryes, supporting our previous findings based on RAMP (Random amplified microsatellite poly-morphic DNA) markers (Shang et al. 2004). For the 13 markers from Saal and Wricke (1999) the average PIC value was 0.589 for 30Secalerepresentatives and 0.383 for cultivated rye, whereas for the 11 markers from Hackauf and Wehling (2002) the average PIC value was 0.620 for 30
Figure 1- The genetic relationships inSecalespecies based on micro-satellite markers.
Table 4- Genetic similarities (GS) and range of variations between cultivated rye accessions from different continents.
Continent Asia Europe North America South America
Asia 0.769 (0.667 to 0.857)
Europe 0.777 (0.634 to 0.902) 0.794 (0.658 to 0.920)
North America 0.744 (0.636 to 0.833) 0.742 (0.622 to 0.867) 0.731 (0.683 to 0.775)
Secalerepresentatives and 0.576 for 47 cultivated ryes, re-spectively. Saal and Wricke (1999) developed the micro-satellite markers from a genomic library, whereas the markers developed by Hackauf and Wehling (2002) were derived from expressed sequence tags (EST) sequences. Our results suggest that the EST-derived microsatellite markers were superior to the microsatellite markers from genomic library, and could reveal a relatively higher level of polymorphism in the genusSecale, especially in culti-vated rye.
The phylogenetic relationships ofSecaleat the spe-cies level has been investigated using diverse techniques (Venseset al. 1987a, Reddyet al. 1990, Del Pozo et al. 1995, De Bustos and Jouve 2002). Our study showed that theS. sylvestreaccessions were very divergent from the
ac-cessions of the other species investigated, agreeing with the majority of previous studies (Reddyet al. 1990, Del Pozoet al. 1995, De Bustos and Jouve 2002). OurS. strictum acces-sions were heterogeneous, with large differences between subspecies. For example, we found that S. strictumssp. africanum, once considered to be a separate species (Khush 1962), was indeed clustered in a separate group, whileS. strictumssp.kuprijonovii, the hypothetical ancestor of the other taxa (Hammer, 1990), was clearly similar to S. strictumssp. Anotolicumbut divergent from the other S. strictum subspecies, supporting the results published by Hammer (1990) and De Bustos and Jouve (2002). Tradi-tionally,S. vaviloviihas been considered to be a separate species (Khush 1962, Del Pozoet al. 1995, Cuadrado and Jouve 1997, De Bustos and Jouve 2002) but we found that it was closely related toS. cereale. No obvious differences were found between theS. cerealesubspecies, supporting the results of SecalerDNA ITS analysis reported by De Bustos and Jouve (2002).
It is thought that cultivated rye originated in the Mount Ararat and Lake Van area of eastern Turkey, lin-guistic evidence suggesting that the introduction of culti-vated rye to southern and western Europe and Central Asia were independent of each other (Sencer and Hawkes 1980). Khush (1962) concluded that cultivated rye probably en-tered Europe by two routes, one being through the northern Caucuses and the other through central Asia. Bushuk (1976) proposed that cultivated rye was probably distrib-uted from south-western Asia to Russia, and thence into Poland and Germany from where it gradually spread throughout most of Europe and eventually to North Amer-ica and western South AmerAmer-ica. Rye was introduced into China from Turkey and later the species was into Japan. Ma et al. (2004) found that American cultivars were more closely related to Chinese cultivars than to European cul-tivars and that temporal isolation had influenced the genetic diversity of rye more than geographical isolation.
In their book concerning the origin of isolating mech-anisms in flowing plants, Maxet al. (1978) observed that geographical, ecological and reproductive isolation should be taken into account when studying plant evolution. In our study we analyzed the genetic similarities of cultivated rye accessions from Asia, Europe, North America and South America, but could not make any deductions regarding the domestication process of cultivated rye, indicating that fur-ther studies are needed to detect the phylogenetic relation-ships and evolution process of cultivated rye.
Acknowledgments
This work was supported by the National High Tech-nology Research and Development Program of China (863 program 2003AA207100) and the Foundation for the Au-thor of National Excellent Doctoral Dissertation of China (200357). Dr. Y.-M. Wei was supported by the Program for New Century Excellent Talents in University of China.
Prof. Y.-L. Zheng was supported by Program for Chang-jiang Scholars and Innovative Research Team in University of China (IRT0453).
References
Anderson JA, Churchill GA, Autrique JE, Tanksley SD and Sor-rells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181-186.
Bushuk W (1976) Rye: Production, Chemistry, and Technology. In: Walter Bushuk (ed). Am Ass Cereal Chemists Inc, St Paul, pp 1-11.
Cuadrado A and Jouve N (1997) Distribution of highly repeated DNA sequences in species of the genus Secale. Genome 40:309-317.
Cuadrado A and Jouve N (2002) Evolutionary trends of different repetitive DNA sequences during speciation in the genus
Secale. J Hered 93:339-345.
De Bustos A and Jouve N (2002) Phylogenetic relationshios of the genusSecalebased on the characterization of rDNA ITS se-quences. Pl Syst Evol 235:147-154.
Delipavlov D (1962)Secale rhodopaeumDelipavlov - A new spe-cies of rye from the Rhodope Mountains. Dokl Bulg Akad Nauk 15:407-411.
Del Pozo JC, Figueiras AM, Benito C and De La Pena A (1995) PCR derived molecular markers and phylogenetic relation-ships in theSecalegenus. Biologia Plant 37:481-489. Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure
from small quantities of fresh leaf tissues. Phytochem Bull 19:11-15.
Frederiksen S and Peterson G (1998) A taxonomy revision of
Secale(Triticeae, Poceae). Nordic J Bot 18:399-420. Hackauf B and Wehling P (2002) Identification of microsatellite
polymorphisms in an expressed portion of the rye genome. Plant Breed 121:17-25.
Hammer K, Skolimowska E and Knüpffer H (1987) Vorarbeiten zur monographischen Darstellung von Wildpfanzensorti-menten:SecaleL. Kulturpflanze 35:135-177.
Hammer K (1990) Breeding system and phylogenetic relation-ships inSecale. Biol Zentralbl 109:45-50.
Khush GS (1962) Cytogenetic and evolutionary studies inSecale. II. Interrelationships of the wild species. Evolution 16:484-496.
Litt M and Luty JA (1989) A hypervariable microsatellite re-vealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle acting gene. Am J Hum Genet 44:397-401.
Ma R, Yli-Mattila T and Pulli S (2004) Phylogenetic relationships among genotypes of worldwide collection of spring and
winter ryes (Secale cerealeL.) determined by RAPD-PCR markers. Hereditas 140:210-221.
Max KH, William CS and Bruce W (1978) The origin of isolating mechanism in flowing plants. In: Max KH (ed) Evolutionary Biology. Plenum Press, New York, pp 185-317.
Nei M and Li W (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269-5273.
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingley S and Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225-238.
Reddy P, Appels R and Baum BR (1990) Ribosomal DNA spacer-length variation in Secale spp.(Poaceae). Pl Syst Evol 171:203-220.
Rohlf FJ (2000) NTSYS-PC Numerical Taxonomy and Multi-variate Analysis System. Version 2.1. Exeter Software, New York.
Roshevitz RY (1947) A monograph of the wildweedy and culti-vated species of rye. Acta Inst Bot Nomine Acad Sci USSR Ser 1. Fe Et Syst 6:105-163.
Saal B and Wricke G (1999) Development of simple sequence re-peat markers in rye (Secale cerealeL.). Genome 42:964-972.
Sencer HA and Haekes JG (1980) On the origin of cultivated rye. Biol J Linn Soc 13:299-313.
Shang HY, Zheng YL, Wei YM, Wu W and Yan ZH (2004) Ge-netic diversity and relationships amongSecaleL. based on RAMP markers. Chin J Agri Biotech 1:73-78.
Tautz D, Trice M and Dover GA (1986) Cryptic simplicity in DNA is a major source of genetic variation. Nature 322:652-656.
Tautz D (1989) Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res 17:6463-6471.
Takezake N and Nei M (1996) Genetic distances and reconstruc-tion of phylogenetic trees from microsatellite DNA. Genet-ics 144:389-399.
Vences FJ, Vaquero F and Perez de la Vega M (1987a) Phloge-netic relationships in Secale: An isozymatic study. Plant Syst Evol 157:33-47.
Vences FJ, Vaquero F, Garcia P and Perez de la Vega M (1987b) Further studies on phylogenetic relationships inSecale: On the origin of its species. Plant Breed 98:281-291.
Zhebentyayeva TN, Reighard GL, Gorina VM and Abbott AG (2003) Simple sequence repeat (SSR) analysis for asseement of genetic variability in apricot germplasm. Theor Appl Genet 106:435-444.