Integration of the Mobilizable
Salmonella
Genomic
Island 1
Benoıˆt Doublet1*, George R. Golding2, Michael R. Mulvey2, Axel Cloeckaert1
1INRA, UR1282, Infectiologie Animale et Sante´ Publique, Nouzilly, France,2National Microbiology Laboratory, Public Health Agency of Canada, Winnipeg, Manitoba, Canada
Abstract
Background:TheSalmonellagenomic island 1 is an integrative mobilizable element (IME) originally identified in epidemic
multidrug-resistantSalmonella entericaserovar Typhimurium (S. Typhimurium) DT104. SGI1 contains a complex integron, which confers various multidrug resistance phenotypes due to its genetic plasticity. Previous studies have shown that SGI1 integrates site-specifically into theS. enterica,Escherichia coli, orProteus mirabilischromosome at the 39end ofthdFgene (attBsite).
Methodology/Principal Findings:Here, we report the transfer of SGI1 to aDthdFmutant ofS.Typhimurium LT2. In the
absence ofthdF, the frequency of transconjugant formation was reduced by around thirty times of magnitude. Through DNA sequencing SGI1 was shown to integrate specifically into a secondary attachment site (2ndattB), which is located in the intergenic region between the chromosomalsodBandpurRgenes. At this 2ndattBsite, we found that a significant fraction of SGI1 transconjugants (43% of wild type and 100% ofDthdFmutant) contained tandem SGI1 arrays. Moreover, in wild type S.Typhimurium LT2 transconjugants, SGI1 integrated into both attachment sites, i.e.,thdFandsodB-purR. The formation of SGI1 tandem arrays occurred in both specificattB sites. There was heterogeneity in the size of the SGI1 tandem arrays detected in single transconjugant colonies. Some arrays consisted as far as six SGI1s arranged in tandem. These tandem arrays were shown to persist during serial passages with or without antibiotic selection pressure.
Conclusions/Significance:The ability of integration into two distinct chromosomal sites and tandem array formation of
SGI1 could contribute to its spread and persistence. The existence of a secondary attachment site in the Salmonella chromosome has potential implications for the mobility of SGI1, which may integrate in other attachment sites of other bacterial pathogens that do not possess the 1stor 2ndspecific SGI1attBsites ofSalmonella.
Citation:Doublet B, Golding GR, Mulvey MR, Cloeckaert A (2008) Secondary Chromosomal Attachment Site and Tandem Integration of the Mobilizable SalmonellaGenomic Island 1. PLoS ONE 3(4): e2060. doi:10.1371/journal.pone.0002060
Editor:Cecile Fairhead, Pasteur Institute, France
ReceivedFebruary 12, 2008;AcceptedMarch 16, 2008;PublishedApril 30, 2008
Copyright:ß2008 Doublet et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding:This research was supported by public funds from the French National Institute of Agronomic Research. Competing Interests:The authors have declared that no competing interests exist.
* E-mail: Benoit.Doublet@tours.inra.fr
Introduction
Genomic islands are large chromosomal regions that have been acquired by horizontal transfer. They are present in certain bacteria but are absent in most closely related bacteria [1]. Genomic islands often carry genes that bring a selective advantage to the host bacterium in a specific environment. Thus, they are classified into pathogenicity islands which encode virulence determinants, resistance islands which confer multiple antibiotic resistances, xenobiotic degradation islands, and symbiosis islands [1,2]. They are frequently integrated near or into tRNA genes, flanked by repeat structures, and contain mobility genes coding for integrases or transposases [1]. However, the majority of genomic islands seem to have lost the ability of horizontal transfer. Burrus et al. proposed to classify as integrative and conjugative elements (ICEs), mobile elements which excise from the chromosome by a site-specific recombination, leading to the formation of circular extrachromosomal element; this intermediate is transferred by conjugation and integrates often in a site-specific fashion into the
recipient chromosome [3]. The genomic islands are widespread in
c-proteobacteria, however few genomic islands have been characterized as mobile elements [3].
The 43-kbSalmonella Genomic island 1 (SGI1) is aS. enterica-derived resistance island that was originally identified in epidemic multidrug-resistant S. enterica serovar Typhimurium phage type DT104 strains [4,5]. The SGI1 contains an antibiotic resistance gene cluster conferring resistance to ampicillin (Ap), chloram-phenicol (Cm), florfenicol (Ff), streptomycin (Sm), spectinomycin (Sp), sulfonamides (Su), and tetracycline (Tc). The 13-kb SGI1 antibiotic resistance gene cluster is located near the 39end of SGI1 and constitutes a complex class 1 integron that belongs to the In4 group, which has been recently named In104 [6,7]. The In104 integron possesses two cassette attachment sites (attI1). At the first attI1site of this complex integron, the cassette carries the aadA2 gene, which confers resistance to Sm and Sp, and downstream a 39
down-stream the 39-CS comprises a complete sul1 gene conferring resistance to Su. Flanked by the two cassettes are thefloRgene, which confers cross-resistance to Cm and Ff, and the tetracycline resistance genestetRandtet(G) . Since the identification of SGI1 in S.Typhimurium DT104, variant SGI1 antibiotic resistance gene clusters have been described in a wide variety ofS. entericaserovars such as Agona, Albany, Cerro, Derby, Dusseldorf, Emek, Infantis, Kentucky, Kiambu, Meleagridis, Newport, and Paratyphi B [5]. Recently, SGI1 and variants of it have been identified inProteus mirabilisclinical and food isolates [5,8–10]. SGI1 variant antibiotic resistance gene clusters were accordingly classified in SGI1-A to SGI1-O [5,10–12]. The identification of SGI1 in P. mirabilis clinical isolates is of great concern as the spread of the SGI1 multidrug resistance phenotype could have significant clinical implications in pathogenic bacteria other thanSalmonella. Potential attachment sites have been identified in diverse human pathogenic bacteria such asShigellaspp.,Vibriospp.,Pseudomonasspp.,Brucella spp.,Legionella pneumophila, andKlebsiella pneumoniaehighlighting the potential for SGI1 to emerge in other human pathogens [9].
In 2005, we reported that SGI1 could be conjugally transferred fromS. entericadonor strains to non-SGI1S. entericaandEscherichia coli recipient strains where it integrated into the recipient chromosome in a site-specific manner [13]. Excision of SGI1 from the Salmonella chromosome occurs through specific recom-bination between the 18-bp direct repeats DR-L and DR-R, mediated by the SGI1-encoded integrase geneint. After excision, the circular extrachromosomal form of SGI1 harbours a unique 18 bp attachment site (attP). After conjugative mobilization intrans, the chromosomal integration of SGI1 occurs via a site-specific recombination between the circular form of SGI1 (attP) and the specific site at the 39 end ofthdFgene (hereafter named primary attBsite) in the recipientS. entericaandE. colichromosome. SGI1 appeared to be a non-self-transmissible but mobilizable element and was thus classified within the group of integrative mobilizable elements (IMEs) that are related to ICEs [13,14].
In the present study, we report the transfer of SGI1 by conjugation to aS.Typhimurium LT2 recipient strain lacking the chromosomalthdFgene, i.e. the primary SGI1attBsite (1stattB). In the absence ofthdF, we found that SGI1 transfer resulted in the integration of SGI1 in a unique secondary integration site (2nd attB) showing conserved regions with the 1st attB site. The integration of SGI1 in its 2nd attB site always resulted in the formation of extended tandem arrays. These tandem arrays had variable copy numbers of SGI1 in the population of single transconjugants. Our findings suggest that the capacity of multiple site integration and tandem SGI1 arrays may contribute to the spread and persistence of multidrug resistance conferred by SGI1.
Results
Conjugative transfer of SGI1 in the absence of thethdF gene inS. Typhimurium LT2 recipient strain
To examine whether SGI1 integration is limited to its 1stattBsite, i.e. the last 18 bp ofthdF, and whether integration in secondary attachment sites may occur, we constructed aDthdFdeletion mutant ofS. Typhimurium strain LT2 whose genome sequence is available (GenBank accesion number NC_003197) (Table 1) [15]. We realized mating experiments using SGI1-F carryingS.Albany strain 7205.00 as donor strain [16] which harbours different somatic O antigens compared to rifampicin-resistant wild type orDthdFmutant S. Typhimurium LT2 recipient strains. As previously described, SGI1 is not self-transmissible and requires additional conjugative functions provided intransby a helper plasmid [13]. Therefore, we introduced the conjugative helper plasmid R55 in theS. Albany
SGI1 donor strain 7205.00. In the presence of the donor strain,S. Albany 7205.00, harbouring the R55 plasmid, SGI1 transconjugants were obtained using the wild type orDthdFmutantS. Typhimurium LT2 recipient strains. The frequency of transconjugants formation was approximately thirty times reduced in the absence of thdF (Table 2). Wild type or DthdF mutant S. Typhimurium LT2 transconjugants showed the antibiotic resistance profile conferred by SGI1-F (ApCmFfSuTcTm) [16]. The serovar of transconjugants (Typhimurium) was also confirmed by somatic O antigens agglutination tests and specific PCRs for the retron sequence downstream thethdFgene which has been only described in serovar Typhimurium (data not shown). The presence of SGI1 in transconjugants was also confirmed by a set of PCR mappings of the island (antibiotic resistance gene cluster, SGI1 integrase geneint) (data not shown) [16]. Since SGI1 is not able to replicate autonomously, theDthdFmutantS. Typhimurium LT2 transconju-gants recovered in these experiments likely carried SGI1 integrated in alternative chromosomal attachment sites.
SGI1 integrates into a unique secondary integration site
To assess where the integration of SGI1 occurred in the chromosome of the DthdF mutant S. Typhimurium LT2 transconjugants, we examined the left SGI1 junctions in the chromosome for three different transconjugants by ligation-mediated PCR as described in the Materials and Methods section. The SGI1 integration in these transconjugants was determined by sequencing the junctions between the left end of SGI1 and the chromosome. The resulting DNA sequences were then compared to the complete genome sequence of S. Typhimurium LT2 (GenBank accession number NC_003197) [15]. Interestingly, by ligation-mediated PCR two PCR products of 550 bp and 900 bp were obtained for each transconjugant tested (data not shown). The sequence of the first one of 550 bp corresponded to the 59end of SGI1 linked to the 39end separated by the SGI1attPsite of 18 bp. This result suggested a potential tandem integration of SGI1 (see below). The sequence of the second 900 bp PCR product corresponded to the 59junction in the chromosome. In the three transconjugants, SGI1 was found integrated in the intergenic region between the chromosomal genessodBand purR(Fig. 1B). SGI1 was thus integrated downstream of thesodBgene coding for the iron superoxide dismutase and 208 bp upstream of thepurR gene coding for the transcriptional repressor for purine nucleotide synthesis (Fig. 1B). According to the annotated genome sequence of S.Typhimurium LT2, the integration of SGI1 would not be predicted to affect the promoter-operator region ofpurR. PCR was performed using primers FwsodB-RvintSGI1 and FwS044-RvpurR corresponding respectively to the left and right junctions of SGI1 integrated betweensodBandpurRin theS.Typhimurium LT2 chromosome. Ten out of 10 different DthdF mutant S. Typhimurium LT2 SGI1 transconjugants were positive for the left junction between the 59 end of SGI1 (int gene) and the chromosomalsodBgene (data not shown). For the right junction, PCR results were positive between the 39end of SGI1 (S044) and thepurRgene of theS.Typhimurium LT2 chromosome.
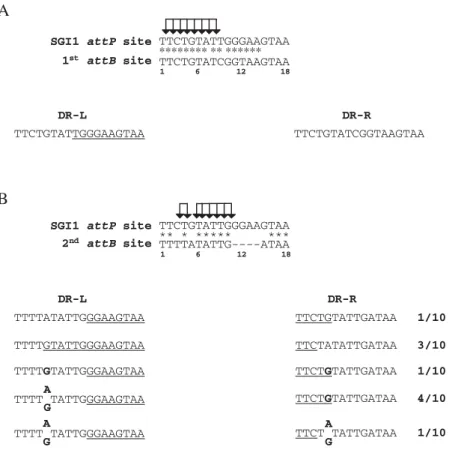
presents two nucleotide substitutions at positions 9 and 12 (Fig. 2A). Analysis of DR-L and DR-R inS.Typhimurium DT104 in which SGI1 is integrated in its 1stattBsite demonstrated that these two nucleotide substitutions were always found in the DR-R (Fig. 2A). This result suggests that the cleavage and strand exchange occur somewhere upstream the position 9 during SGI1 integration in its 1st attBsite (Fig. 2A). The integration of SGI1 in the 2ndattBsite was slightly different. For the 10DthdFmutantS. Typhimurium LT2 transconjugants, the sequences of the left and right junctions were determined to analyze the direct repeat sequences flanking SGI1 in
this 2ndattBsite (Fig. 2B). As shown in Figure 2B, the sequence of the 2ndattBsite in theS.Typhimurium LT2 chromosome differs both in length and sequence from the specific SGI1 attP sequence. Compared to the SGI1attPsite, the 2ndattBsite is only 14 bp in length and presents three additional substitutions at positions 3, 5, and 15 to the four gap positions (Fig. 2B). The differences in the SGI1attPsite and the 2ndattBsite result in different DR-L and DR-R sequences that allow the cleavage sites during recombination betweenattPandattBto be estimated (Fig. 2B). The sequences of DR-L and DR-R suggest that one cleavage and DNA strand
Table 2.Effect of theDthdF::kanmutation ofS.Typhimurium LT2 recipient strain on the SGI1 transfer frequency.
SGI1 donor strain Conjugative helper plasmid R55 Recipient strain SGI1 transfer frequencya
S.Albany 7205.00 2 Wild typeS.Typhimurium LT2 ,1029
S.Albany 7205.00 2 DthdFmutantS.Typhimurium LT2 ,1029
S.Albany 7205.00 + Wild typeS.Typhimurium LT2 2.1 1024
S.Albany 7205.00 + DthdFmutantS.Typhimurium LT2 7.6 1026
athe frequency of transfer was caculated by dividing the number of SGI1 transconjugants by the number of SGI1 donor cells. Transfer frequencies correspond to the means of three independent mating experiments.
doi:10.1371/journal.pone.0002060.t002
Table 1.Bacterial strains, plasmids, and primers used in this study.
Strains and plasmid Relevant genotype and resistance profileaor characteristics Reference or source
S.enterica
Albany 7205.00 SGI1-F+
; ApCmFfSuTcTm [16]
Typhimurium LT2 Sensitive, sequenced genome [15]
Typhimurium LT2 SGI12; Rif This study
Typhimurium LT2DthdF::kan SGI12; RifKm This study
Plasmids
IncC R55 (K. pneumoniae) Tra+
; ApCmFfGmKmSu [19]
pKD4 Derivative pANTSc, containing an FRT-flanked kanamycin resistance (kan); ApKm [38] pKD46 Derivative pINT-ts,lRed recombinase under control ofParaBpromoter; Ap [38] Primersb
U7-L12 ACACCTTGAGCAGGGCAAAG [4]
LJ-R1 AGTTCTAAAGGTTCGTAGTCG [4]
104-RJ TGACGAGCTGAAGCGAATTG [4]
C9-L2 AGCAAGTGTGCGTAATTTGG [4]
104-D ACCAGGGCAAAACTACACAG [4]
RecthdF-F AGGCGGTCATATGACCGCCTTTTTTTATTGCAACAAAGTTGAGACTAACCGTGTAGGCTGGAGCTGCTTC This study RecthdF-R TTACGGGTTTTGTAGGCCCGGTAAGCATCGTGCCACCGGGCAACACAACGCATATGAATATCCTCCTTAG This study
Linker1 TAATTACACGTTACGACTTCAGATC This study
Linker2 GATCTGAAGTCGTAACGTG This study
RvintLM TTCTTTATTGTGCTGACGCTCTG This study
SGI1circ1 AGCAAAATCGTGAGAAGGGA [13]
SGI1circ2 TGATGAGACACCTGACGAGC [13]
FwsodB GAAAAATCTCGCCGCATAAG This study
RvintSGI1 CCTCACCTTCAACAACTCCG This study
FwS044 CTACCCAGGAGCCACAATCA This study
RvpurR GCCCGTTTCGCTACATCTTT This study
aabbreviations: Ap, ampicillin; Cm, chloramphenicol; Ff, florfenicol; Gm, gentamicin; Km, kanamycin; Rif, rifampicin; Su, sulphonamides; Tc, tetracyclines; Tm, trimethoprim
bNucleotide sequences are indicated from 5
exchange occur between bases 3 and 5 (3 out of 10) of the core sequence and the other cleavage and strand exchange occur somewhere between bases 5 and 11 (1 out of 10) (Fig. 2B). Interestingly, in one transconjugant a G nucleotide was found at position 5 in both DR-L and DR-R. In five other transconjugants, a mix of A and G nucleotides was found at position 5 in DR-L or both in DR-L and DR-R (Fig. 2B). The finding of the same base (G) at position 5 in both DR-L and DR-R could be consistent with mismatch repair of single bp substitutions during recombination. Such event has been previously described for the lambda bacteriophage [17]. Furthermore, the mix of bases (A or G) at position 5 in DR-L or both in DR-L and DR-R suggests that in one transconjugant different subpopulations may present different DR-Ls or different DR-DR-Ls and DR-Rs. This finding is consistent with the hypothesis of tandem integration of SGI1 and then different excision events with cleavage and strand exchange upstream or downstream position 5. Thus, SGI1 excision events in tandem arrays could generate for one transconjugant different subpopulations with different DR-Ls or different DR-Ls and DR-Rs.
Transfer of SGI1 promotes SGI1 tandem arrays in recipient strains
To assess whether SGI1 integration occurred in tandem arrays inS.Typhimurium LT2 recipient strains, we examined the SGI1 junctions for wild type andDthdF mutantS. Typhimurium LT2 transconjugants by Southern blot hybridization. A 364-bp SGI1 attPprobe containing part of S044, the 18 bpattPsite and part of theintgene was used. The whole genomic DNAs of 6 wild typeS. Typhimurium LT2 transconjugants digested by HindIII were probed with this SGI1attPprobe (Fig. 3A). For all transconjugants, this probe revealed twoHindIII fragments (Fig. 3A) of the expected sizes (Fig. 1A) corresponding to the left and right junctions when SGI1 is integrated in its 1stattBsite, i.e., the last 18bp ofthdF. Four of 6 of the transconjugants studied had a 2.8-kbHindIII SGI1 attP-specific fragment which corresponded to the link between the 39end and 59end of SGI1 (Figs. 1A, 3A). Six DthdF mutant S. Typhimurium LT2 transconjugants were also studied by Southern blot hybridization using this probe and the restriction enzymeBglI according to the sequence surrounding the 2ndattBsite (Figs. 1B,
B B
5553 bp
B B B B B B
6691 bp 5553 bp 3466 bp
SGI1
thdF yidY
retron
SGI1
sodB SGI1 SGI1 purR
DR-L TTCTGTATTGGGAAGTAA
DR-R
TTTT TATTGGGAAGTAA TTCT TATTGATAA
attP TTCTGTATTGGGAAGTAA
attP TTCTGTATTGGGAAGTAA A
G
A G
SGI1 tandem arrays
SGI1 tandem arrays
thdF yidY
retron
1stattB
Primary integration site
A
sodB purR
2ndattB TTTTATATTGATAA Secondary integration site
2ndDR-L 2ndDR-R
TTCTGTATCGGTAAGTAA TTCTGTATCGGTAAGTAA
B
attP TTCTGTATTGGGAAGTAA
SGI1
H H H H H H
1017 bp 2844 bp 2428 bp
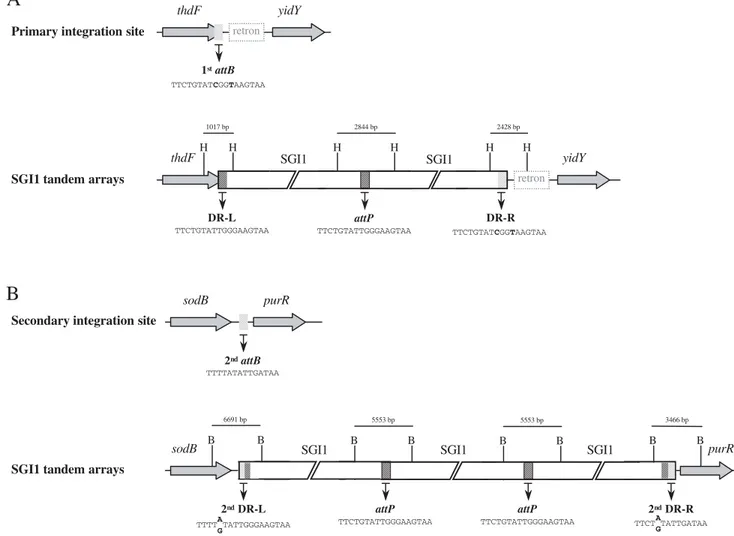
Figure 1. Chromosomal integration sites and tandem arrays of SGI1 in theS.Typhimurium chromosome.(A)Schematic view of the
primary specific integration site (1stattB) of SGI1 within the 3
9end of the chromosomalthdFgene of differentS.Typhimurium strains. The retron sequence downstream the 1stattBsite is only present inS.Typhimurium strains. The integration of SGI1 tandem arrays (2 copies) in its 1stattBsite is represented. Positions ofHindIII restriction sites (H) are indicated without respect of bp scale to show the expected sizes ofHindIII fragments corresponding to 1stDR-L, 1stDR-R, andattP.(B)Schematic view of the secondary integration site (2ndattB) of SGI1 between the chromosomalsodB andpurRgenes. The integration of SGI1 tandem arrays (3 copies) in the 2ndattBsite is represented. Positions ofBglI restriction sites (B) are indicated without respect of bp scale to show the expected sizes ofBglI fragments corresponding to 2ndDR-L, 2ndDR-R, andattP. The sequences of 1stattB, 2nd attB,attP, DR-Ls, and DR-Rs are indicated.
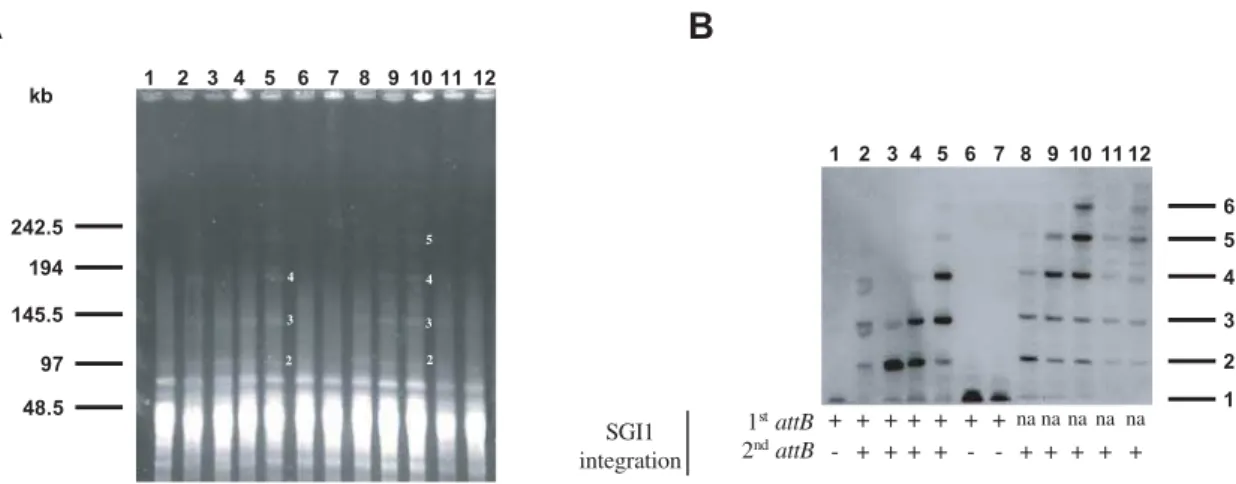
3B). TheattPprobe revealed threeBglI fragments of the expected sizes corresponding to 2ndDR-L, 2ndDR-R, and the 5.5 kbBglI attP-specific fragment (Figs. 1B, 3B). The specific-attP fragment revealed in Figure 3A and 3B could be derived from circular extrachromosomal SGI1 or from chromosomal integrated tandem arrays of SGI1. The first possibility appeared unlikely, as in differentSalmonellafield strains carrying SGI1, we were unable to extract a circular intermediate of SGI1 by different alkaline lysis extraction methods and moreover the detection of this circular form by PCR required a nested PCR [13]. Therefore, we used pulsed-field gel electrophoresis (PFGE) to assess whether the specific attP fragment revealed by Southern blot hybridization represented integrated copies of SGI1 arranged in tandem or not. To demonstrate SGI1 tandem arrays in PFGE, we used the restriction enzyme AscI which does not cut within SGI1 and frequently cut the S. Typhimurium LT2 chromosome in small fragments (around 10 kb in size). According to the genome sequence ofS.Typhimurium LT2 and the 42,433 bp size of SGI1, the expected sizes of one SGI1 copy integrated at its 1stor 2ndattB sites are 51 and 56 kb, respectively. In this manner, the expected sizes of different tandem arrays were determined. DNA fromS. Albany strain 7205.00, which contained a single SGI1 copy was used as control and 6 wild type and 5 DthdF mutant S. Typhimurium LT2 transconjugants were tested. Compared to theAscI restriction patterns ofS.Albany strain 7205.00, new bands
of higher molecular weight appeared in both the AscI digested DNAs of wild type and DthdF mutant S. Typhimurium LT2 transconjugants (Fig. 4A). To conclude on the copy number of SGI1 arranged in tandem and to exclude the possibility of large AscI chromosomal fragments, we hybridized the PFGE gel with a specific SGI1 probe (p1-9 probe [6]) (Fig. 4B). This Southern blot hybridization revealed six different large fragments of expected sizes consistent with the presence of one, two, three, four, five, and six SGI1s integrated in tandem in the chromosome. TheS.Albany control strain and 2 out of 6 wild type transconjugants presented a single integrated SGI1 copy (Fig. 4B). For these transconjugants, the integration of SGI1 in its 1stattBsite and its absence in its 2nd attBsite was confirmed by PCRs using primers U7L12-LJR1 and FwsodB-RvpurR, respectively (Table 1, Fig. 4B). Interestingly, different SGI1 copy numbers in tandem arrays were found for the four remaining wild type transconjugants and the sixDthdFmutant S. Typhimurium LT2 transconjugants. Thus, these results indicated that different subpopulations resulting from a single transconjugant colony contained different copy number tandem arrays of SGI1. These results are in accordance with the given hypothesis on DR-L and DR-R sequence analysis ofDthdFmutant S. Typhimurium LT2 transconjugants (Fig. 2B). All these transconjugants were tested for the left and right junctions of SGI1 with the chromosome by PCR to confirm the integration site of SGI1 (Fig. 4B). The 6 DthdF mutant S. Typhimurium LT2 TTCTGTATTGGGAAGTAA
TTTTATATTG----ATAA
2nd attB site
SGI1 attP site
TTTTATATTGGGAAGTAA TTCTGTATTGATAA
TTTTGTATTGGGAAGTAA TTCTATATTGATAA
TTCTGTATTGATAA
TTTT TATTGGGAAGTAA
A G
1/10 3/10
4/10
TTTTGTATTGGGAAGTAA TTCTGTATTGATAA 1/10
TTTT TATTGGGAAGTAA
A
G 1/10
TTCT TATTGATAAA
G
** * ***** ***
DR-L DR-R
TTCTGTATTGGGAAGTAA
TTCTGTATCGGTAAGTAA
1st attB site
SGI1 attP site
TTCTGTATTGGGAAGTAA TTCTGTATCGGTAAGTAA
******** ** ******
DR-L DR-R
A
B
18
1 6 12
18
1 6 12
Figure 2. Comparison of the SGI1 18 bpattP,attB, DR-Ls, DR-Rs ofS.Typhimurium.(A)Alignment of theattPsite of SGI1 and the primary attBsite (1stattB) ofS.Typhimurium strain LT2. The sequence of direct repeats left (DR-L) and right (DR-R) flanking integrated SGI1 were indicated.(B) Alignment of theattPsite of SGI1 and the secondaryattBsite (2ndattB) ofS.Typhimurium strain LT2. The sequence of direct repeats left (DR-L) and right (DR-R) were determined in ten independentDthdFmutantS.Typhimurium LT2 SGI1 transconjugants from three mating experiments. (*) indicated identical positions in theattPsite and theattBsites. Positions 1, 6, 12, and 18 are indicated below the 1stand 2ndattBsequences. The specific nucleotides forattPare underlined in DR-Ls and DR-Rs. Sites of possible cleavage during the strand exchange are indicated by arrows inattP. Nucleotides in boldface letters represent illegitimate base pair in the DR-L and DR-R sequences. The sequences of DR-L and DR-R of some transconjugants revealed a mix of G and A at position 5 in repeated attempts.
transconjugants were positive for the SGI1 integration in its secondary attB site and harboured SGI1 tandem arrays. Interestingly, the 4 wild type transconjugants harbouring SGI1 tandem arrays were positive for integration both in the 1stand 2nd attBsites. These results indicated that SGI1 was able to integrate in two distinctattBsites in a single wild typeS.Typhimurium LT2 transconjugant. Thus, the great heterogeneity in subpopulations of SGI1 transconjugants seemed to concern the copy number of tandemly arranged SGI1 and also the integration site.
Simultaneous integration in two specific chromosomal sites and stability of SGI1 tandem arrays
To study whether integration of SGI1 in wild type S. Typhimurium LT2 transconjugants always occurred in the 1st attBsite, or if SGI1 tandem array formation was correlated with one or bothattBsites, we further studied the formation of SGI1 tandem arrays and the integration attB sites using PCR. One hundred SGI1 transconjugants from three independent mating DR-L: 1017 bp
DR-R: 2428 bp
attP: 2844 bp
DR-L: 6691 bp
DR-R: 3466 bp
attP: 5553 bp
1 2 3 4 5 6
A
HindIIIB
BglI
Figure 3. Chromosomal junctions of tandemly-arranged SGI1 islands in theS.Typhimurium LT2 chromosome.(A)Southern blot
hybridization with the 364-bpattPSGI1 probe containing part of S044, 18 bpattPsite and part of theintgene ofHindIII-digested genomic DNAs of wild typeS.Typhimurium LT2 SGI1 transconjugants. Lanes 1-4 correspond to different transconjugants with tandem arrays of SGI1 integrated at the 39end ofthdFand lanes 5-6 to transconjugants with a single SGI1 integrated at thisattBsite.(B)Southern blot hybridization with the 364-bpattP SGI1 probe ofBglI-digested genomic DNA ofDthdFmutantS.Typhimurium LT2 SGI1 transconjugants. All transconjugants tested showed the same profile withBglI fragments containing DR-L, DR-R, andattP.The molecular sizes ofHindIII orBglI fragments containing DR-L, DR-R, andattPare indicated.
doi:10.1371/journal.pone.0002060.g003
242.5
194
145.5
97
48.5
1 2 3 4 5 6 7 8 9 10 11 12
1 2 3 4 5 6 7 8 9 10 11 12 kb
5
4
3
2
1 6
A
B
SGI1 integration
1stattB 2ndattB
+
-+ +
+ + + +
+ +
+
-+
-na
+
na
+
na
+
na
+
na
+
2 3 4 5
2 3 4
Figure 4. Copy numbers of SGI1 tandem arrays in theS. Typhimurium LT2 chromosome. (A)Macrorestriction analysis by PFGE of
genomic DNAs cut byAscI ofS.Typhimurium LT2 SGI1 transconjugants. Lanes: 1,S.Albany strain 7205.00 (SGI1 donor strain); 2-7,S.Typhimurium LT2 SGI1 transconjugants; 8-12,DthdFmutantS.Typhimurium LT2 SGI1 transconjugants. The molecular sizes in kilobases are indicated to the left of the panel. The numbers indicated to the right of the bands on the restriction patterns (lane 5 and 10) correspond to the copy numbers of SGI1 in each fragment. The size of the AscI fragments observed are consistent with the expected sizes of 2 to 5 SGI1 copies in tandem.(B)Southern blot hybrization with the p1-9 probe of the PFGE-AscI restriction patterns of Figure 4A. The numbers indicated to the right of the panel correspond to the copy numbers of SGI1 in each fragment revealed by the p1-9 probe. The integration sites of SGI1 are indicated under each transconjugant profiles; (+) integrated SGI1, (2) unoccupiedattBsite, (na) not applicable.
experiments for the wild type andDthdFmutantS. Typhimurium LT2 recipient strains were tested by PCR junctions for the 1stand 2ndattBsites and by PCR for tandem arrays (junction between two copies of SGI1). The frequencies of site integration and tandem array formation are indicated as percent in Table 3. For all wild type S. Typhimurium LT2 transconjugants, SGI1 was found integrated in its 1st attB site. Forty-three percent of these conjugants also possessed SGI1 integrated in the 2nd attB site.
Interestingly, the same forty-three transconjugants were positive for SGI1 tandem arrays (Table 3). For the one hundred DthdF mutant S. Typhimurium LT2 transconjugants, they were all positive for integration in the 2nd attB site and tandem array
formation (Table 3). These results indicated that the SGI1 integration occurred preferentially in its 1st attB site. However, approximately half of transconjugants harboured integrated SGI1 copies in the two specificattBsites. Moreover, the integration of SGI1 in bothattBsites seemed to be correlated to tandem array formation. Interestingly, the formation of SGI1 tandem arrays always occurred into its 2ndpreferentialattBsite inDthdFmutantS. Typhimurium LT2 transconjugants (in absence of the 1stattBsite). The SXT element fromVibrio choleraewas also able to integrate in a tandem fashion inE. coliinto its specific integration site [18]. However, after 5 days cultures only one copy number of SXT was detected in theE. colitransconjugant suggesting a decrease from a multiple copy number arranged in tandem to only one after this time [18]. To investigate the stability of SGI1 tandem arrays inS. Typhimurium LT2, wild type andDthdFmutant transconjugants were cultivated for 15 days with two dilutions per day in fresh medium (approximately .600 generations) with or without antibiotic selection for SGI1. Wild type transconjugants with only one copy of SGI1 or with tandem arrays were included in this experiment. Throughout this time, bacterial cultures were tested by PCR for SGI1 tandem arrays and at the end time for integration intoattBsites. With or without antibiotic selection, no changes were observed by PCRs for integration sites, single SGI1 copy or tandem arrays during this time (data not shown). Unlike theVibrio choleraeSXT element, SGI1 tandem arrays appeared to persist after several cultures with or without antibiotic selection with at least two SGI1 copies arranged in tandem.
Discussion
Salmonellagenomic island 1 (SGI1) is an integrative mobilizable element (IME) containing an antibiotic resistance gene cluster identified in several S. enterica serovars and recently also in P. mirabilis [5,6,8,10,16]. In a previous study, SGI1 was found to transfer by conjugative mobilization, using conjugative helper plasmid R55 [19], from aS. entericadonor to a recipient strain (E. coli or S. enterica) [13]. In the donor strain, the excision and circularization of SGI1 is mediated by the SGI1-encoded integrase
Int which presents similarity to thel integrase family (Tyrosine recombinase family) [13,20]. The Int-mediated recombination between the 18 bp direct repeats left and right (DR-L and DR-R) flanking the integrated SGI1 results in a unique 18 bp sequence (attPsite) in the SGI1 circular form. The SGI1 integration into the chromosome of the recipient occurs by recombination between the SGI1attPsite of the circular form and the chromosomal 1stattB site, i.e., the last 18 bp of the thdF gene [13]. The site-specific integration of SGI1 in the chromosome demonstrated experimen-tally, is also supported by the growing number ofS. entericaserovars andP. mirabilisstrains in which SGI1 was found to be integrated at the 39end of the chromosomalthdFgene [5,8–10]. Thus, SGI1 represents a non replicative element which needs to integrate in the chromosome to persist in the host strain [4,6,13]. In this study, in absence of the thdF gene, SGI1 was found to integrate in a specific 2nd attB site between the chromosomal sodB and purR genes. However, in some transconjugants containingthdF, SGI1 was found integrated in the two attachment sites,thdFand sodB-purR,with at least one copy in each attachment site. Moreover, tandem arrays of SGI1 were always found in S.Typhimurium LT2 SGI1 transconjugants lackingthdF. There was heterogeneity in the size of SGI1 tandem arrays detected in cells from single transconjugants. Tandem arrays contained different copy numbers of SGI1 ranging in size from two to six repeats.
Various elements, including phages, integrative conjugative elements and pathogenicity islands have been described to integrate site-specifically in one site and also in secondary attachment sites [18,21–26]. Other mobile elements such as the SRL PAI ofShigella flexneri, the clc element ofPseudomonassp. strain B13, and the SXT element of V. choleraeshare with SGI1 very similar properties of integration [3,22,26–28]. The 66-kb SRL (Shigellaresistance locus) PAI (pathogenicity island) inShigellaspp. mediates multiple antibiotic resistances and integrates site-specifically into two bacterial tRNA attB sites [22,27]. The integrase Int of SRL PAI mediates the integration adjacent to one or both identical paralogous tRNA genes serX and serW [22,27]. Chromosomal integrations of the 105-kb clc element of Pseudomonassp. strain B13 occurred also in two similar sites which are the glycine tRNA genes in thePseudomonaschromosome [28]. The SRL PAI island and the clc element are able to integrate in one or both identicalattBsites of the host chromosome. The SXT element ofV. Choleraeis a conjugative self-transmissible chromo-somally integrating element which also contains several antibiotic resistance genes [3,29]. SXT integrates site-specifically at the 59
end of the chromosomalprfCgene [26]. In the absence ofprfC, the SXT element integrates in several secondary attachment sites but preferentially into the 59end of the chromosomalpntBgene [26]. Moreover, the SXT element is also able to integrate in a tandem fashion after conjugative transfer [18].
Table 3.Integration sites and tandem arrays of SGI1 inS.Typhimurium strain LT2.
S.Typhimurium LT2 transconjugant genotype Integration inattBsites (%)a SGI1 tandem arrays (%)c
Primary site 39endthdF Secondary sitesodB-purR
Wild typeS.Typhimurium LT2 100 43 43
DthdFmutantS.Typhimurium LT2 nab 100 100
aThe percent of integration at each site was determined by PCR junctions with the chromosome on one hundred wild type or
DthdFmutantS.Typhimurium LT2 transconjugants from three independent mating experiments.
bNot Applicable.
The mechanism accounting for the formation of SGI1 tandem arrays is unknown but this event is likely related to the conjugative transfer of SGI1. To date in S. enterica field strains harbouring SGI1, we have never detected SGI1 arrays (data not shown). Serial passage of representative field strains containing a single SGI1 did not result in amplification of the SGI1 copy number (data not shown). Several conjugative-dependent mechanisms could explain the formation of tandem SGI1 arrays. Tandem arrays could form if a concatemer of several SGI1 copies was transferred from a single donor cell to a single recipient cell. SGI1 being a mobilizable element, its conjugative transfer is similar to conjugative plasmids transfer. We hypothesize that a single-stranded SGI1 generated by a rolling circle process, is transferred from donor to recipient. The general model of bacterial conjugation proposes that a single strand is transmitted in the 59
to 39orientation to the recipient cell [30]. This transfer process is initiated by nicking DNA and finished by religation at the origin of transfer (oriT) resulting in a monomeric circle of transferred DNA. During the transfer, synthesis of the replacement strand by a rolling circle mode of DNA replication reconstitutes the transferred single strand [30]. Thus, concatemer of several copies could be transferred to the recipient. Alternatively, a single donor may transfer a single SGI1 but in repeated attempts to a single recipient cell. Another explanation is that a single recipient cell could be implicated in successive conjugation events with different donor cells and thus acquired several SGI1 copies.
Interestingly, we demonstrated that the SGI1 integration in the 2ndattBsite always occurred in a tandem fashion in absence of the 1st attB site (Table 3). This result is consistent with a SGI1 concatemer integration in the 2ndattBsite. Moreover, according to the lower frequency of transfer in the DthdF mutant S. Typhimurium LT2 recipient strains, this hypothesis could be more probable than successive transfers of single SGI1 monomer into a single recipient cell. However, the formation of SGI1 tandem arrays also occurred in the wild typeS.Typhimurium LT2 recipient strain. Fifty-seven of one hundred transconjugants tested, presented a single SGI1 integrated in the 1st attB site (Table 3, Fig. 4B). In contrast, for fourty-three transconjugants positive for tandem arrays, integration of SGI1 occurred into the twoattBsites (Table 3). This result suggests that tandem array formation could occur in bothattBsites. Thus, some wild type transconjugants may contain SGI1 tandem arrays integrated in the 1stattBsite (Figs. 3, 4) and one or several SGI1 copies integrated in the 2ndattBsite. DNA fingerprint analysis using PFGE in Figure 4 of wild typeS. Typhimurium LT2 transconjugants did not permit a conclusion as to where SGI1 tandem arrays were integrated between the 1stand 2ndattBsites (only 5 kb size difference). However, the absence of detectable hybridized fragments corresponding to the 2ndDR-L and 2ndDR-R in Southern blot hybridization using the SGI1attP
probe (Fig. 3A) for the four transconjugants containing tandem arrays suggested that the SGI1 tandem arrays were integrated in the 1stattBsite.
Several questions remain to be answered: (i) were there two independent conjugative transfers resulting in the occupancy of the twoattBsites; or (ii) was there in a first time a SGI1 concatemer transfer and integration in the 1stattBsite and then excision of one
or several SGI1 copies and reintegration in the 2ndattBsite in a subpopulation of a single transconjugant. To assess these hypotheses, further studies need to be undertaken to demonstrate a potential sequential event. For thestx2bacteriophages of Shiga
toxin-producingE. coli, the insertion site occupancy by stx phages depends on the availability of the preferred site in the host strain [21]. If the primary insertion site is unavailable, then a secondary insertion site is selected [21]. In contrast, the integrative
conjugative element SXT fromV. choleraeis able to use the left or right direct repeats of a previously integrated element for integration [31].
The analysis of SGI1 copy number in tandem arrays in transconjugants revealed considerable heterogeneity (Fig. 4). Although formation of tandem SGI1 arrays appears to occur frequently inS.Typhimurium LT2 chromosome after conjugative transfer in vitro, these arrays probably decreased in size during bacterial multiplication resulting in different subpopulations with different SGI1 copy numbers in a single colony. Homologous recombination between SGI1 copies in a tandem array may lead to a decreased SGI1 copy number in arrays. Another explanation is that the SGI1 integrase Int could excise single or multiple SGI1s by recombination involving the DR-L and an internalattPsite of the tandem arrays, or DR-R and an internal attP site, or two internal attP sites. All these recombinations would result in the formation of circular extrachromosomal forms containing a single or several SGI1s in the bacteria. According to the previous speculation, such circular intermediates could be implicated in integration in the remainingattBsite. Moreover, analysis of DR-L and DR-R inDthdFmutantS.Typhimurium LT2 transconjugants (Fig. 2B) demonstrated different DR-L and/or DR-R sequences in some transconjugants with mix of bases at position 5. This result suggested that integration or excision of SGI1 copies occurred at the left and/or right side of tandem arrays in the 2ndattBsite.
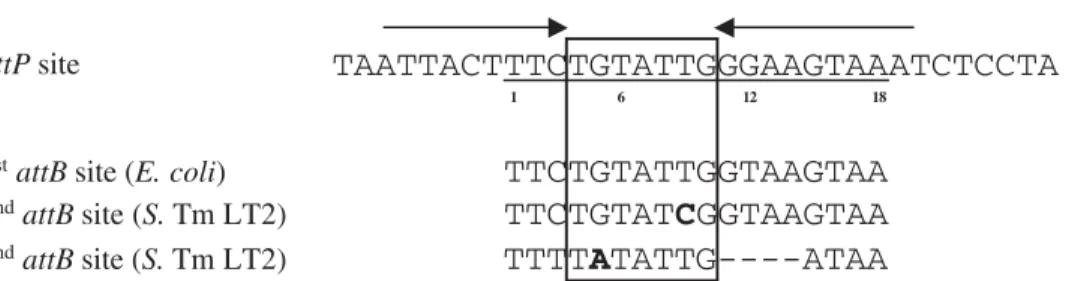
strongly dependent on the presence of homology between the first 3 bp of overlap regions [20,35]. The lower transfer frequency usingDthdFmutantS. Typhimurium LT2 recipient compared to wild type could be an indication for a lower integration frequency in the 2ndattBsite potentially due to the substitution at the position 5 in the 7-bp overlap region (Fig. 5). However, other integrase-binding sites like arm type sites or core-type sites are also described to play an important role in integration frequency [20]. Site-directed mutagenesis could be used to establish which positions within this putative 7-bp overlap region ofattBsites are critical for the integration of SGI1.
In summary, we have shown that the genomic island SGI1 is able to integrate in a secondaryattBsite which is highly conserved amongst the different Salmonella sequenced genomes (data not shown). After conjugative transfer, SGI1 tandem arrays are integrated in bothattBsites with a great heterogeneity in the size of the tandem arrays in single transconjugant colonies. The ability of integration into distinct chromosomal sites could contribute to the spread and persistence of SGI1. Thus, SGI1 could possibly integrate in other bacterial pathogens that do not possess either the 1st or 2ndSGI1 attB sites but a slightly divergent attB site. It is interesting to note that several genomic islands implicated in multidrug resistance are now described to use site-specific integration in the host chromosome as a mean for persistence after horizontal transfer. This study provides an interesting insight into potential mechanisms that strengthen the spread of multiple antibiotic resistance among human bacterial pathogens.
Materials and Methods
Bacterial strains, plasmids and antibiotic susceptibility testing
The Salmonella strains used in conjugation experiments are described in Table 1. S. Albany strain 7205.00 harbouring the SGI1-F variant was used as donor strain [16]. S.Typhimurium strain LT2 was made rifampicin resistant as previously described [15,36]. All strains were grown at 37uC in brain heart infusion broth or agar plates. IncC conjugative plasmid R55 fromKlebsiella pneumoniae was used as a helper plasmid for mobilization experiments as previously described [13,19]. Donor, recipient, and transconjugant strains were screened for antibiotic resistance by the disk diffusion method on Mueller-Hinton agar plates [37]. Susceptibility was tested using disks containing the following antibiotics: Ap (10mg), Cm (30mg), Ff (30mg), Km (30 IU), Gm (15mg), Sm (10 IU), Sp (100mg), Su (200mg), Tc (30 IU) and trimethoprim (Tm) (5 mg). All antibiotic disks except Ff were purchased from BioRad (Marnes la Coquette, France). Ff disks were obtained from Schering-Plough Animal Health (Segre´, France).
Deletion of thethdF gene by insertion mutagenesis
Deletion of the chromosomal thdFgene was performed in S. Typhimurium strain LT2 using the one step chromosomal gene inactivation technique [38]. Briefly, the kanamycin resistance gene kanflanked by FRT (FLP recognition target) sites was amplified by standard PCR using the template plasmid pKD4 and hybrid primers. These primers, RecthdF-F and RecthdF-R (Table 1), were synthesized with 20 nucleotides of priming sites of pKD4 and with 50 nucleotides from each side of thethdFgene. The 1.6 kb long PCR fragment was purified and electroporated into the S. Typhimurium strain LT2 in which the l Red recombinase expression plasmid pKD46 was introduced. Homologous recom-bination between the genomic DNA and the PCR product resulted in the deletion of the entire thdF gene and in its replacement with the kangene. The resulting strain was named
DthdFmutantS.Typhimurium LT2 compared to the wild typeS. Typhimurium strain LT2.
Bacterial conjugations
Conjugation assays were performed by mixingS.Albany SGI1 donor strain 7205.00 with or without the helper plasmid R55 and the rifampicin resistant S. Typhimurium LT2 recipient strains (wild type or DthdF mutant) together with a donor-to-recipient ratio of 4:1. This broth was incubated overnight at 37uC without shaking. The next day, the cells were streaked on appropriate selective brain heart infusion agar plates. Rifampicin (250mg/ml) was used to select againstS.Albany donor cells, and Tc (10mg/ml) to select against unmated recipient cells. The SGI1 frequency of transfer was determined by dividing the number of SGI1 transconjugants by the number of S. Albany SGI1 donor cells. Transconjugants were tested for antibiotic resistance, for somatic O antigens by agglutination tests with antisera (Bio-Rad, Manes la Coquette, France), and also by PCR for specific markers described below.
Secondary attachment site determination by ligation-mediated PCR
The secondary integration sites of SGI1 were determined by performing ligation-mediated PCR as described below. Genomic DNAs ofDthdFmutant S.Typhimurium LT2 SGI1 transconju-gants were cut by blunt-end restriction enzymes AluI or EcoRV (Promega, Charbonnieres, France). Annealing of the two primers Linker1 and Linker2 to form the double-stranded adaptators was performed by boiling a 5 nM solution of the mixed primers, followed by slow cooling to room temperature. AluI or EcoRV digested chromosomal DNAs were ligated to adaptators in 10ml final volume at a 10-fold molar excess of the adaptator, according to the number of generated fragments.
TAATTACTTTCTGTATTGGGAAGTAAATCTCCTA
TTCTGTAT
C
GGTAAGTAA
TTTT
A
TATTG----ATAA
attP
site
2
ndattB
site (
S.
Tm LT2)
2
ndattB
site (
S.
Tm LT2)
TTCTGTATTGGTAAGTAA
1
stattB
site (
E. coli
)
18
1 6 12
Figure 5. Analysis of the SGI1attPoverlapping region.The previously described 18-bp sequence of theattPsite is underlined. Positions 1, 6, 12, and 18 are indicated below theattPsite. The imperfect inverted repeats are indicated by arrows. Sequences of the different SGI1attBsites are aligned below the SGI1attPsequence. Nucleotide in boldface letters represented substitutions compared to theattPsequence. The black box represented the putative 7-bp overlap region in which cleavages and strand exchanges occurred.
A first round of amplification was performed by using the primer Linker1 and the first SGI1 internal primer RvintLM to the left end of SGI1 (Table 1), in 25ml PCR mixtures with a GoTaq Master Mix kit (Promega, Charbonnieres, France) and 2ml of ligation. The first-round PCR conditions were (i) 5 min at 95uC, (ii) 30 cycles of 30 s at 95uC, 30 s at 60uC, and a variable elongation time at 72uC according to the length of generated fragments, and (iii) 7 min at 72uC. The second round of amplification was performed like the first round with 2ml of the first-round reaction mixture as the template and primers Linker1 and LJR1, which was identical to the leftmost end of SGI1 (Table 1). The second-round amplification conditions were (i) 5 min at 95uC, (ii) 30 cycles of 30 s at 95uC, 30 s at 57uC, and variable elongation time at 72uC, and (iii) 7 min at 72uC. The purified PCR products were sequenced by using the SGI1 LJR1 primer at Genome Express (Meylan, France) and were compared with the GenBank DNA sequence database by using the genomic BLASTN program.
PCR mapping, sequencing, Southern blot hybridization
Detection of SGI1 and its location were performed using primers corresponding to the left and right junction in the 1stand 2nd attB integration site (Table 1, Fig. 1). PCR products corresponding to the left and right junctions at the secondary attBsite of ten independentDthdFmutantS.Typhimurium LT2 transconjugants were sequenced. Nucleotide sequencing was achieved by Genome Express (Meylan, France). For the wild type and DthdF mutant S. Typhimurium LT2 recipient strains, one hundred independent transconjugants of three different mating experiments were screened by PCR on the left junction for SGI1 integration (1st and 2nd attB sites) and by PCR using primers SGI1circ1 and SGI1circ2 oriented towards the left and right end of SGI1 for tandem integration (Table 1, Fig. 1).
To assess tandem arrays of SGI1, Southern blot analysis of wild type andDthdFmutantS.Typhimurium LT2 transconjugants was performed. Briefly, total genomic DNAs of transconjugants were digested with HindIII or BglI and hybridized with the 364-bp amplified fragment containing part of S044,attP, and part of SGI1 as a probe. The expected sizes of fragments containing L, DR-R, andattPin wild type andDthdF::kantransconjugants correspond to 1017, 2428, 2844 bpHindIII fragments and 6691, 3466, 5513 bpBglI fragments, respectively.
Copy number of SGI1 in tandem arrays
Chromosomal DNA of wild type and DthdF mutant S. Typhimurium LT2 SGI1 transconjugants strains was prepared for pulsed field gel electrophoresis as previously described [16]. Genomic DNA was digested with AscI restriction enzyme (BioLabs, Saint Quentin, France), which do not cut within SGI1 but relatively frequently in the chromosome ofS.Typhimurium LT2. Fragments of DNA were separated by pulsed-field gel electrophoresis (PFGE) in a 1% agarose gel (BioRad, Marnes la Coquette, France) by using a CHEF-DR III (Bio-Rad, Hemel Hempstead, United Kingdom). The running conditions were 6 V/ cm at 14uC for 22 h, with pulse times ramped from 7 to 20 s. Southern blot hybridization was realized onAscI PFGE using the p1-9 probe previously described to assess the copy number of SGI1 in tandem arrays.
Acknowledgments
We thank C. Mouline and K. Praud for expert technical assistance.
Author Contributions
Conceived and designed the experiments: BD AC. Performed the experiments: BD. Analyzed the data: BD GG MM AC. Contributed reagents/materials/analysis tools: BD GG. Wrote the paper: BD MM AC.
References
1. Dobrindt U, Hochhut B, Hentschel U, Hacker J (2004) Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2: 414–424. 2. Burrus V, Pavlovic G, Decaris B, Gue´don G (2002) Conjugative transposons: the
tip of the iceberg. Mol Microbiol 46: 601–610.
3. Burrus V, Waldor MK (2004) Shaping bacterial genomes with integrative and conjugative elements Shaping bacterial genomes with integrative and con-jugative elements. Res Microbiol 155: 376–386.
4. Boyd D, Peters GA, Cloeckaert A, Sidi Boumedine K, Chaslus-Dancla E, et al. (2001) Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region ofSalmonella entericaserovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183: 5725–5732.
5. Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A (2006) The genetics ofSalmonellagenomic island 1. Microbes Infect 8: 1915–1922.
6. Boyd D, Cloeckaert A, Chaslus-Dancla E, Mulvey MR (2002) Characterization of variantSalmonellagenomic island 1 multidrug resistance regions from serovars Typhimurium DT104 and Agona. Antimicrob Agents Chemother 46: 1714–1722. 7. Levings RS, Lighfoot D, Partridge SR, Hall RM, Djordjevic SP (2005) The genomic island SGI1, containing the multiple resistance region ofSalmonella entericaserovar Typhimurium DT104 or variants of it, widely distributed in other S. entericaserovars. J Bacteriol 187: 4401–4409.
8. Ahmed AM, Hussein AIA, Shimamoto T (2007)Proteus mirabilisclinical isolate harbouring a new variant ofSalmonellagenomic island 1 containing the multiple antibiotic resistance region. J Antimicrob Chemother 59: 184–190.
9. Doublet B, Golding GR, Mulvey MR, Cloeckaert A (2007) Potential integration sites of theSalmonellagenomic island 1 inProteus mirabilisand other bacteria. J Antimicrob Chemother 59: 801–803.
10. Boyd DA, Shi X, Hu QH, Ng LK, Doublet B, et al.SalmonellaGenomic Island 1 (SGI1), Variant SGI1-I, and a New Variant SGI1-O inProteus mirabilisClinical and Food Isolates from China. Antimicrob Agents Chemother 52: 340–344. 11. Cloeckaert A, Praud K, Doublet B, Demartin M, Weill FX (2006) Variant
Salmonellagenomic island 1-L antibiotic resistance gene cluster inSalmonella entericaserovar Newport. Antimicrob Agents Chemother 50: 3944–3946. 12. Vo ATT, van Duijkeren E, Fluit AC, Gaastra W (2007) A novelSalmonella
genomic island 1 and rare integron types inSalmonellaTyphimurium isolates from horses in The Netherlands. J Antimicrob Chemother 59: 594–599.
13. Doublet B, Boyd D, Mulvey MR, Cloeckaert A (2005) The Salmonella genomic island 1 is an integrative mobilizable element. Mol Microbiol 55: 1911–1924.
14. Burrus V, Pavlovic G, Decaris B, Gue´don G (2002) The ICESt1element of Streptococcus thermophilusbelongs to a large family of integrative and conjugative elements that exchange modules and change their specificity of integration. Plasmid 48: 77–97.
15. McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, et al. (2001) Complete genome sequence ofSalmonella entericaserovar Typhimurium LT2. Nature 413: 852–856.
16. Doublet B, Lailler R, Meunier D, Brisabois A, Boyd D, et al. (2003) Variant Salmonellagenomic island 1 antibiotic resistance gene cluster inSalmonella enterica serovar Albany. Emerg Infect Dis 9: 585–591.
17. Bauer CE, Gardner JF, Gumport RI, Weisberg RA (1989) The effect of attachment site mutations on strand exchange in bacteriophagelsite-specific recombination. Genetics 122: 77–735.
18. Burrus V, Waldor MK (2004) Formation of SXT tandem arrays and SXT-R91 Hybrids. J Bacteriol 186: 2636–2645.
19. Gaffney DF, Cundliffe E, Foster TJ (1981) Chloramphenicol resistance that does not involve chloramphenicol acetyltransferase encoded by plasmids from Gram negative bacteria. J Gen Microbiol 125: 113–121.
20. Azaro MA, Landy A (2002)lintegrase and thelInt family. In: Craig NL, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA II. Wahington: ASM press. pp 118–148.
21. Serra-Moreno R, Jofre J, Muniesa M (2007) Insertion site occupancy bystx2
bacteriophages depends on the locus availibility of the host strain chromosome. J Bacteriol 189: 6645–6654.
22. Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B (2004) Role ofattPin integrase mediated integration of theShigellaresistance locus pathogenicity island ofShigella flexneri. Antimicrob Agents Chemother 48: 1028–1031.
23. Rutkai E, Dorgai L, Sirot R, Yagil E, Weisberg RA (2003) Analysis of insertion into secondary attachment sites by phageland byintmutants with altered recombination specificity. J Mol Biol 329: 983–996.
25. Song B, Shoemaker NB, Gardner JF, Salyers AA (2007) Integration site selection by theBacteroidesconjugative transposon CTnBST. J Bacteriol 189: 6594–6601. 26. Burrus V, Waldor MK (2003) Control of the SXT integration and excision.
J Bacteriol 185: 5045–5054.
27. Turner SA, Luck SN, Sakellaris H, Rajakumar K, Adler B (2003) Molecular epidemiology of the SRL pathogenicity island ofShigella flexneri2a. Antimicrob Agents Chemother 47: 727–734.
28. Ravtan R, Studer S, Springael D, Zehnder AJB, van der Meer JR (1998) Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes fromPseudomonassp. strain B13. J Bacteriol 180: 4360–4369.
29. Hochhut B, Waldor MK (1999) Site-specific integration of the conjugalVibrio choleraeSXT element intoprfC. Mol Microbiol 32: 99–110.
30. Wilkins B, Lanka E (1993) DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell DB editor. Bacterial conjugation? New-York: Plenum Press. pp 105–136.
31. Hochhut B, Beaber JW, Woodgate R, Waldor MK (2001) Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J Bacteriol 183: 1124–1132.
32. Zucker M (2003) Mfold web server for nucleic acid folding and hbridization prediction. Nucleic Acids Res 31: 3406–3415.
33. Malanowska K, Salyers AA, Gardner LF (2006) Characterization of a conjugative transposon integrase IntDOT. Mol Mic 60: 1228–1240. 34. Weeslund NA, Wang GR, Song B, Shoemaker NB, Salyers AA (2007)
Integration and excision of a newly discoveredBacteroidesconjugative transposon, CTnBST. J Bacteriol 189: 1072–1082.
35. Burgin AB, Nash HA (1995) Suicide substrates reveal properties of th homology-dependent steps during integrative recombination of bacteriophagel. Curr Biol 5: 1312–1321.
36. Golding GR, Olson AB, Doublet B, Cloeckaert A, Christianson S, et al. (2007) The effect of theSalmonellagenomic island 1 on in vitro global gene expression in Salmonella entericaserovar Typhimurium LT2. Microbes Infect 9: 21–27. 37. Members of the SFM antibiogramm committee (2003) comite´ de
l’antibio-gramme de la socie´te´ franc¸aise de microbiologie report 2003. Int J Antimicrob Agents 21: 364–391.