Circulating MicroRNAs as a Novel Class of Diagnostic
Biomarkers in Gastrointestinal Tumors Detection: A
Meta-Analysis Based on 42 Articles
Ran Wang1,2, Hong Wen2, Yongcheng Xu3, Qiulan Chen2, Yi Luo2, Yiqin Lin2, Yu Luo2, Angao Xu1,3* 1Department of Gastroenterology, Nanfang Hospital, Southern Medical University, Guangzhou, China,2Department of Ultrasound, HuiZhou Municipal Central Hospital, Huizhou, China,3Department of Gastroenterology, HuiZhou Municipal Central Hospital, Huizhou, China
Abstract
Objective:MicroRNAs (miRNAs) have become the focus of most recent efforts in cancer research. However, there have been inconsistencies in the literature regarding the suitability of circulating miRNAs for early detection of gastrointestinal cancers. This study aims to assess the diagnostic performance of circulating miRNAs in detection of gastrointestinal cancer through a meta-analysis.
Methods:Eligible studies were selected by conducting a systematic literature search of public databases. The sensitivity and specificity were used to plot the summary receiver operator characteristic (SROC) curve and calculate the area under the SROC curve (AUC). The between-study heterogeneity was evaluated byQtest andI2statistics. Subgroup analyses and meta-regression were further performed to explore the potential sources of heterogeneity. All analyses were performed using the STATA 12.0 software.
Results:A total of 107 studies from 42 articles were included for the meta-analysis according to the inclusion criteria. The overall analysis of all gastrointestinal cancers showed that circulating miRNAs have a relatively good diagnostic performance in gastrointestinal cancers, with a sensitivity of 0.75, a specificity of 0.81 and an AUC of 0.85. In addition, subgroup analyses based on different type of miRNA assay suggested that single-miRNA assay displayed a relatively low diagnostic performance with the AUC values of 0.84 for gastric cancer (GC) and 0.79 for colorectal cancer (CRC), while multiple-miRNAs assay significantly improved the diagnosing accuracy with AUC rising to 0.92 for GC and 0.89 for CRC. Another interesting finding was that plasma-based miRNA assay reach a higher accuracy compared with serum-based one for GC, while opposite conclusion was drawn for CRC.
Conclusions:In conclusion, circulating miRNAs, particularly the combination of multiple miRNAs, may present as promising biomarkers for the diagnosis of gastrointestinal cancers. Further large-scale prospective studies are necessary to validate their potential applicability in human cancer diagnosis.
Citation:Wang R, Wen H, Xu Y, Chen Q, Luo Y, et al. (2014) Circulating MicroRNAs as a Novel Class of Diagnostic Biomarkers in Gastrointestinal Tumors Detection: A Meta-Analysis Based on 42 Articles. PLoS ONE 9(11): e113401. doi:10.1371/journal.pone.0113401
Editor:Olga Y. Gorlova, Geisel School of Medicine at Dartmouth College, United States of America ReceivedJune 19, 2014;AcceptedOctober 23, 2014;PublishedNovember 18, 2014
Copyright:ß2014 Wang et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data Availability:The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are contained within the paper and references.
Funding:The authors have no support or funding to support.
Competing Interests:The authors have declared that no competing interests exist. * Email: angao_xu@163.com
Introduction
Gastrointestinal tract cancers, especially gastric, colorectal, and esophageal cancers, are one of the most common causes of cancer-related deaths [1]. It was estimated that tumors in esophagus, stomach, and colorectum account for approximately 11% of all newly diagnosed cancers and 14% of cancer related deaths in the United States in 2013, which make it an epidemiological health concern [2]. Currently, one of the biggest challenges in cancer treatment is the lack of specific and sensitive biomarker for early cancer diagnosis, which hinders the patients from receiving the timely treatment. The 5-year survival rate after surgical resection reaches 90% for gastric cancer (GC) patients at stage I, but this rate dramatically drops to 5% in cases at stage IV [3]. For
colorectal cancer (CRC) patients, the 5-year survival rate of stage II cases is over 80% after surgical resection, but less than 10% at advanced [4]. In addition, the locoregional recurrence and/or distant metastasis can be frequently observed in the late-stage cancer patients even if they have already received the resection and multimodality therapy [5]. Therefore, the low survival rate of cancer patients at advanced stages highlights the importance of early cancer diagnosis. Unfortunately, most human cancers show no symptom in at early stages, which makes it hard for early diagnosis, and the cost-effectiveness of available diagnostic techniques is unsatisfactory.
Currently, the wide range of conventional diagnostic methods, including gastroscopy, random biopsies, colonoscopy, double contrast barium enema (DCBE), and computed tomographic
colonography (CTC), are applied to diagnose and monitor gastrointestinal cancers. Although gastroscopy/colonoscopy is currently considered to be the most reliable screening tool with reportedly high accuracy, its invasive nature and expensive cost have hindered its widespread application in cancer diagnosis as a screening tool [6,7]. DCBE and CTC can detect some intestinal cancers at an early stage, but the complicated diagnostic procedures as well as the associated radiation hazards also limit their clinical applications [8]. In addition, fecal-based analyses, such as occult blood and stool DNA tests, are currently most common non-invasive procedure for early cancer diagnosis [9,10]. However, the lack of sufficient sensitivity and specificity hampers their utility in the detection of premalignant lesions.
It is generally believed that cancer-related biomarkers in blood would be quite helpful in early cancer diagnosis and tumor progression monitoring. Several currently available circulating biomarkers, such as carbohydrate antigen 19-9 (CA 19-9), carcinoembryonic antigen (CEA), pepsinogen (PG) I/II ratio, estrogen receptor (ER), and progesterone receptor (PR), are being used as non-invasive methods for cancer diagnosis without involving a biopsy or a surgical procedure [11–13]; Unfortunately, they also suffer from the limitation of low sensitivities and specificities. Therefore, there is a pressing need for novel and more sensitive non-invasive biomarkers to improve the diagnostic accuracy for gastrointestinal cancers.
Recently, circulating miRNAs have attracted considerable attention in its diagnostic value for human cancers. MiRNAs are a family of small non-coding functional RNAs with 19–24 nucleotides modulating the expression of messenger RNA (mRNA) [14]. In recent years, miRNAs have been found to be dysregulated in a variety of diseases, particularly in human cancers [15,16]. It has been observed that miRNAs could present extensively in the cell-free body fluids and excretions, including serum, plasma, urine, tears, saliva, bronchial lavage, and feces, etc [17]. Furthermore, biochemical analyses indicate that circulating miRNAs have a remarkable stability and are tolerant to RNase activity and extreme physiological environment [18], making it plausible to use circulating miRNAs as novel non-invasive biomarkers in diagnosing and monitoring human cancers.
The role of miRNAs as novel biomarkers in cancer was first recognized in a study considering miR-15 and miR-16, which were found to be down-regulated in B-cell chronic lymphocytic leukemia (B-CLL) [19]. Subsequent evidence has indicated that unique miRNA expression profiles in circulation may contribute to the diagnosis of cancers, such as colorectal cancer [20], gastric cancer [21], esophageal cancer [22], breast cancer [23], lung cancer [24], hepatocellular carcinoma [25], prostate cancer [26], and pancreatic cancer [27]. However, there have been incon-sistencies or discrepancies in the literature reviews regarding the reliability of circulating miRNAs for early detection of
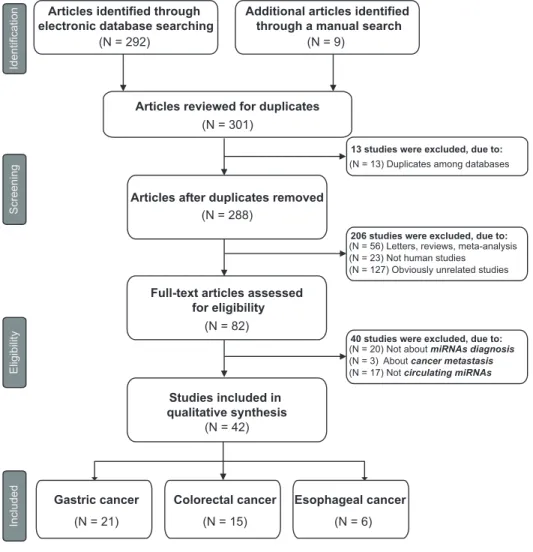
gastro-Figure 1. Flow diagram of study selection process.
doi:10.1371/journal.pone.0113401.g001
Circulating MiRNAs for Gastrointestinal Tumors
Table 1.Main characteristics of 42 studies included in meta-analysis.
Included studies Cancer type Location Ethnicity Study design Case/Control miRNA profiling Specimen QUADAS
Number Mean age Male ratio
Cai H, 2013 GC China Asian Case-control 90/90 46.2/46.1 0.81/0.81 miR-106b, -20a, -221 Plasma 5
Chen Q, 2014 GC China Asian Case-control 36/36 56/59 0.78/0.78 miR-122, -192 Plasma 6
Konishi H, 2012 GC Japan Asian Case-control 56/30 66/NA 0.55/NA miR-451, -486 Plasma 7
Li BS, 2012 GC China Asian Case-control 60/60 54/51 0.70/0.63 miR-223, -21, -218 Plasma 4
Li C, 2013 GC China Asian Case-control 180/80 58.1/58.9 0.31/0.35 miR-199a-3p, -151-5p Plasma 5
Li ZQ, 2012 GC China Asian Case-control 46/21 58.8/57 0.65/0.67 miR-27a, -181b Plasma 5
Liu H, 2012 GC China Asian Case-control 40/41 56/58 0.66/0.66 miR-371-5p, -187, -378 Serum 3
Liu R, 2011 GC China Asian Case-control 164/127 60.2/60 0.84/0.8 miR-1, -20a, -27a, -34, -423-5p
Serum 4
Peng WZ, 2014 GC China Asian Case-control 57/58 NA/43 NA/0.48 miR-191, -425 Serum 5
Sheinerman KS, 2013
GC America Caucasian Case-control 10/30 55.8/51.9 0.10/0.30 miR-203, -146b-5p, -192 Plasma 6
Shiotani A, 2013 GC Japan Asian Case-control 64/64 67.9/68.4 0.70/0.64 miR-106b, let-7 Serum 5
Song MY, 2012 GC China Asian Retrospective 68/68 60.4/60.4 0.71/0.71 7 miRNAsa Serum 4
Tsujiura M, 2010 GC Japan Asian Case-control 69/30 NA/NA NA/NA miR-106b Plasma 5
Valladares M, 2012 GC Spain Caucasian Case-control 52/15 65.9/65.3 0.81/0.81 miR-200c Plasma 7
Wang B, 2012 GC China Asian Case-control 30/39 58/46 0.73/0.77 miR-21 Serum 3
Xu Q, 2013 GC China Asian Case-control 94/103 60.2/59.1 0.63/0.63 miR-320a Serum 4
Zheng Y, 2011 GC China Asian Case-control 53/20 60/60 0.66/0.66 miR-21 Serum 4
Zhou H, 2010 GC China Asian Case-control 90/27 62.3/NA 0.70/NA miR-106a, -17 Serum 4
Zhou H, 2012 GC China Asian Case-control 40/17 64.6/64.6 0.73/0.73 miR-421 Serum 4
Zhu C, 2014 GC China Asian Case-control 40/40 53.8/53.5 0.73/0.73 miR-16, -25, -92a, -451, -468-5p
Plasma 3
Zhu CJ, 2011 GC China Asian Case-control 48/27 61.2/55.4 0.83/0.74 miR-191, -27a Plasma 6
Feng L, 2013 CRC China Asian Case-control 98/50 54.3/52.7 0.53/0.52 miR-92a Serum 7
Giraldez MD, 2013 CRC Spain Caucasian Cohort 42/53 62.8/62.1 0.51/0.49 miR-19a, -19b, -15b Plasma 4
Huang Z, 2010 CRC China Asian Case-control 100/59 61/58 0.51/0.53 miR-29a, -92a Plasma 3
Liu et al, 2013 CRC China Asian Cohort 200/80 57.9/57.5 0.63/0.52 miR-21, 92a Serum 6
Liu HS, 2012 CRC China Asian Case-control 47/28 58.6/56.2 0.66/0.68 miR-129-3p, -767-3p, -877
Serum 4
Luo X, 2013 CRC Germany Caucasian Case-control 80/144 68/62.5 0.56/0.42 12 miRNAsb Plasma 5
Ng EK, 2009 CRC China Asian Case-control 90/50 71/69 0.52/0.52 miR-17-3p, -92 Plasma 5
Pu XX, 2010 CRC China Asian Cohort 103/37 58/32 0.64/0.51 miR-221 Plasma 7
Sheinerman KS, 2013
CRC America Caucasian Case-control 10/30 57.8/57.3 0.80/0.30 miR-203, -146b-5p, -192, -215, -303-3p
Plasma 3
Toiyama Y, 2013 CRC Japan Asian Cohort 186/53 67.5/64 0.57/0.51 miR-21 Serum 3
Circulating
MiRNAs
for
Gastrointestin
al
Tumors
PLOS
ONE
|
www.ploson
e.org
3
November
2014
|
Volume
9
|
Issue
11
|
Table 1.Cont.
Included studies Cancer type Location Ethnicity Study design Case/Control miRNA profiling Specimen QUADAS
Number Mean age Male ratio
Wang Q, 2012 CRC China Asian Case-control 90/58 62/58 0.50/0.52 miR-601, -760, -29a, -92a Plasma 6
Wang S, 2013 CRC China Asian Cohort 77/84 64/44 0.42/0.60 miR-409-3p, -7, -93 Plasma 4
Yong FL, 2013 CRC Malaysia Asian Case-control 70/32 64.4/61.5 0.60/0.48 miR-193a-3p, -23a, -338-5p
Serum 5
Zanutto S, 2014 CRC Italy Caucasian Cohort 29/29 NA/NA NA/NA miR-378, -21 Plasma 5
Zhang GJ, 2013 CRC China Asian Case-control 78/86 61.4/60.3 0.55/0.62 miR-200c, 18a Plasma 4
Hirajima S, 2013 EC Japan Asian Case-control 106/54 65/NA 0.82/NA miR-18a Plasma 5
Komatsu S, 2011 EC Japan Asian Case-control 50/20 65/NA 0.88/NA miR-21/-375 Plasma 7
Takeshita N, 2013 EC Japan Asian Case-control 101/46 NA/NA 0.89/NA miR-1246 Serum 3
Zhang C, 2010 EC China Asian Cohort 149/100 62/50 0.78/0.74 7 miRNAsc Serum 4
Zhang T, 2011 EC China Asian Cohort 201/202 NA/NA 0.66/0.63 miR-31 Serum 3
Zhang T, 2013 EC China Asian Case-control 201/201 NA/NA 0.66/0.62 miR-1322 Serum 6
amiR-221, -744, -376c, -27a, -27b, -222, -191.
bmiR-18a, -20a, -21, -29a, -92a, -106b, -133a, -143, -145, -181b, -342-3p, -532-3p. cmiR-10a, -22, -100, -148b, -223, -133a, -127-3p.
NA, not available; GC, gastric cancer; CRC, colorectal cancer; EC, esophageal cancer; QUADAS-2, the revised Quality Assessment of Diagnostic Accuracy Studies. doi:10.1371/journal.pone.0113401.t001
Circulating
MiRNAs
for
Gastrointestin
al
Tumors
PLOS
ONE
|
www.ploson
e.org
4
November
2014
|
Volume
9
|
Issue
11
|
intestinal cancers. In this meta-analysis, we summarize an overview of circulating miRNAs present in blood circulation to further elucidate their diagnostic performance and provide information for the early detection of gastrointestinal cancers.
Materials and Methods
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidance (Supplement S1).
Literature search
This meta-analysis was conducted according to guidelines for diagnostic meta-analysis [28]. Eligible studies published up to 1 April 2014 were selected for meta-analysis by conducting a systematic literature search of public databases including PubMed, Embase, Chinese National Knowledge Infrastructure (CNKI), and Chinese Biology Medicine (CBM) databases, without language limitation. The following retrieval strategy was used: (‘gastro-intestinal tumor’ OR ‘gastric tumor’ OR ‘gastric cancer’ OR ‘colorectal tumor’ OR ‘colorectal cancer’ OR ‘esophageal tumor’ OR ‘esophageal cancer’) AND (‘microRNA’ OR ‘miRNA’ OR ‘miR’) AND (‘blood’ OR ‘serum’ OR ‘plasma’ or ‘circulating’) AND (‘diagnosis’ OR ‘sensitivity and specificity’ OR ‘ROC curve’). In addition, reference lists of eligible articles were independently searched manually to obtain additional sources.
Inclusion and exclusion criteria
Eligible studies included in this meta-analysis have to fulfill the following criteria: (1) studies regarding the diagnostic potential of circulating miRNAs for gastrointestinal cancers; (2) studies with a gold reference standard for the gastrointestinal cancers diagnosis; (3) studies with sufficient data for construction of two-by-two tables [i.e., true positive (TP), false positive (FP), true negative (TN) and false negative (FN)]. Exclusion criteria were: (1) publications unrelated to the diagnostic values of circulating miRNAs for gastrointestinal cancers; (2) studies with duplicate data reported in other studies; (3) letters, editorials, case reports or reviews.
Data extraction
Two reviewers independently extracted data from all the eligible studies: (1) basic characteristics of studies, including name of the first author, year of publication, country of origin, ethnicity, study design, sample size, mean age, male ratio, cancer type, type of miRNA assay, methods of miRNAs detection, type of specimens; and (2) diagnostic performance, including sensitivity, specificity, TP, FP, FN, and TN.
Quality assessment
The qualities of included studies were scored independently by two reviewers using the revised Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) criteria [29]. The QUADAS-2 tool is comprised of 4 key domains: patient selection, index test, reference standard and flow and timing, and uses seven questions to evaluate the quality of included studies (Supplements S2). Each question is answered with ‘‘yes’’, ‘‘no’’, or ‘‘unclear’’. An answer of ‘‘yes’’ means that the risk of bias can be judged low, while an answer of ‘‘no’’ or ‘‘unclear’’ means that the risk of bias can be judged high. In case of conflict, a third reviewer was consulted, and disagreement was settled through multilateral discussion.
Statistical analysis
All analyses were performed using the STATA 12.0 software. The bivariate meta-analysis model was employed to summarize the sensitivity (SEN), specificity (SPE), positive likelihood ratio (PLR), negative likelihood ratio (NLR) and diagnostic odds ratio (DOR) [30,31]. The sensitivity and specificity of each included study were used to plot the summary receiver operator character-istic (SROC) curve and calculate the area under the SROC curve (AUC). The AUC can be statistically interpreted as the probability to correctly distinguish patients from normal controls. The between-study heterogeneity was evaluated by Q test and I2 statistics. APvalue less than 0.10 forQtest orI2values$50% indicates substantial heterogeneity, and then the random-effects model was applied [32,33]. To further explore the potential sources of heterogeneity, subgroup analyses and meta-regression were performed according to the characteristics of the included studies. As publication bias is a concern for meta-analyses, Deeks’ funnel plot asymmetry test was used, with P,0.10 indicating statistically significant [34].
Results
Procedure of literature retrieval
The procedure of the literature retrieval was presented in Figure 1. The initial search returned a total of 301 articles, of which 13 duplicate publications among databases were removed. After the review of titles and abstracts, 206 articles were excluded: 56 were reviews or letters, 23 were not human studies, and 127 were not related to our research topic, leaving 82 articles available for further full-text review. After careful reading, 40 articles were
Figure 2. Overall quality assessment of included studies using the QUADAS-2 criteria (a: risk of bias; b: applicability).
doi:10.1371/journal.pone.0113401.g002
Circulating MiRNAs for Gastrointestinal Tumors
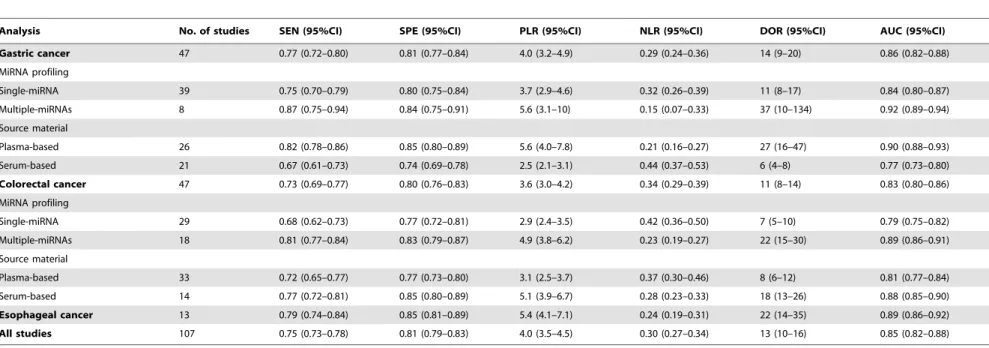
Table 2.Summary estimates of diagnostic criteria and their 95% confidence intervals (95% CI).
Analysis No. of studies SEN (95%CI) SPE (95%CI) PLR (95%CI) NLR (95%CI) DOR (95%CI) AUC (95%CI)
Gastric cancer 47 0.77 (0.72–0.80) 0.81 (0.77–0.84) 4.0 (3.2–4.9) 0.29 (0.24–0.36) 14 (9–20) 0.86 (0.82–0.88)
MiRNA profiling
Single-miRNA 39 0.75 (0.70–0.79) 0.80 (0.75–0.84) 3.7 (2.9–4.6) 0.32 (0.26–0.39) 11 (8–17) 0.84 (0.80–0.87)
Multiple-miRNAs 8 0.87 (0.75–0.94) 0.84 (0.75–0.91) 5.6 (3.1–10) 0.15 (0.07–0.33) 37 (10–134) 0.92 (0.89–0.94)
Source material
Plasma-based 26 0.82 (0.78–0.86) 0.85 (0.80–0.89) 5.6 (4.0–7.8) 0.21 (0.16–0.27) 27 (16–47) 0.90 (0.88–0.93)
Serum-based 21 0.67 (0.61–0.73) 0.74 (0.69–0.78) 2.5 (2.1–3.1) 0.44 (0.37–0.53) 6 (4–8) 0.77 (0.73–0.80)
Colorectal cancer 47 0.73 (0.69–0.77) 0.80 (0.76–0.83) 3.6 (3.0–4.2) 0.34 (0.29–0.39) 11 (8–14) 0.83 (0.80–0.86)
MiRNA profiling
Single-miRNA 29 0.68 (0.62–0.73) 0.77 (0.72–0.81) 2.9 (2.4–3.5) 0.42 (0.36–0.50) 7 (5–10) 0.79 (0.75–0.82)
Multiple-miRNAs 18 0.81 (0.77–0.84) 0.83 (0.79–0.87) 4.9 (3.8–6.2) 0.23 (0.19–0.27) 22 (15–30) 0.89 (0.86–0.91)
Source material
Plasma-based 33 0.72 (0.65–0.77) 0.77 (0.73–0.80) 3.1 (2.5–3.7) 0.37 (0.30–0.46) 8 (6–12) 0.81 (0.77–0.84)
Serum-based 14 0.77 (0.72–0.81) 0.85 (0.80–0.89) 5.1 (3.9–6.7) 0.28 (0.23–0.33) 18 (13–26) 0.88 (0.85–0.90)
Esophageal cancer 13 0.79 (0.74–0.84) 0.85 (0.81–0.89) 5.4 (4.1–7.1) 0.24 (0.19–0.31) 22 (14–35) 0.89 (0.86–0.92)
All studies 107 0.75 (0.73–0.78) 0.81 (0.79–0.83) 4.0 (3.5–4.5) 0.30 (0.27–0.34) 13 (10–16) 0.85 (0.82–0.88)
CI, confidence interval; SEN, sensitivity; SPE, specificity; PLR, positive likelihood ratio; NLR, negative likelihood ratio; DOR, diagnostic odds ratio; AUC, area under the curve. doi:10.1371/journal.pone.0113401.t002
Circulating
MiRNAs
for
Gastrointestin
al
Tumors
PLOS
ONE
|
www.ploson
e.org
6
November
2014
|
Volume
9
|
Issue
11
|
further excluded: 20 were not about gastrointestinal cancers diagnosis, 3 were relevant to metastasis in cancers, and 17 were not circulating miRNAs. Finally, 42 articles were included according to the inclusion criteria, 21 of which focused on GC [21,35–54], 15 on CRC [20,49,55–67], and the other 6 on EC [22,68–72].
Baseline characteristics of included studies
The main characteristics of 42 articles were shown in Table 1. In total, 107 studies from these 42 articles were involved in the current meta-analysis. As for GC, 47 studies from 21 articles were
available for analysis. 38 of these 47 studies investigated the diagnostic value of single-miRNA assay in GC detection, while only 8 focused on multiple-miRNAs assay. 26 studies used plasma as specimen, and the other 21 based on serum. Similarly, for 47 studies from the 15 articles focusing on CRC, 29 of them assessed the performance of single-miRNA assay in CRC detection, whereas 18 focused on multiple-miRNAs assay. 33 studies used plasma as specimen, while serum was applied in the other 14 studies. For EC diagnosis, 13 studies from 6 articles were available. The publication years of the included articles range from 2010 to 2014. All studies used the quantitative real-time reverse
transcrip-Figure 3. SROC curve with pooled estimates of sensitivity, specificity and AUC (a: overall studies on gastrointestinal cancers; b: GC; c: CRC; d: EC).
doi:10.1371/journal.pone.0113401.g003
Circulating MiRNAs for Gastrointestinal Tumors
tion-PCR (qRT-PCR) method to measure the expression of circulating miRNAs. Quality assessment for the overall studies was shown with a bar graph according to the QUADAS-2 tool in Figure 2. The majority of all included studies in this meta-analysis fulfilled 4 or more of the 7 items in QUADAS-2, indicating that the overall quality of included studies is generally good.
Diagnostic accuracy of miRNAs in gastrointestinal cancers
The pooled estimates of gastrointestinal cancers (GC/CRC/ EC) for the diagnostic accuracy of circulating miRNAs were
presented in Table 2. The overall analysis of all types gastro-intestinal cancers showed that circulating miRNAs have a relatively good diagnostic performance in gastrointestinal cancers, with SEN of 0.75 (95% CI: 0.73–0.78), SPE of 0.81 (95% CI: 0.79–0.83) and AUC of 0.85 (95% CI: 0.82–0.88) (Figure 3A). Since only 13 studies from 6 publications were involved in EC, subgroup analyses were conducted for GC and CRC. For EC, the diagnostic accuracy of circulating miRNAs was even better than the overall results, with SEN of 0.79 (95% CI: 0.74–0.84), SPE of 0.85 (95% CI: 0.81–0.89) and AUC of 0.89 (95% CI: 0.86–0.92) (Figure 3D).
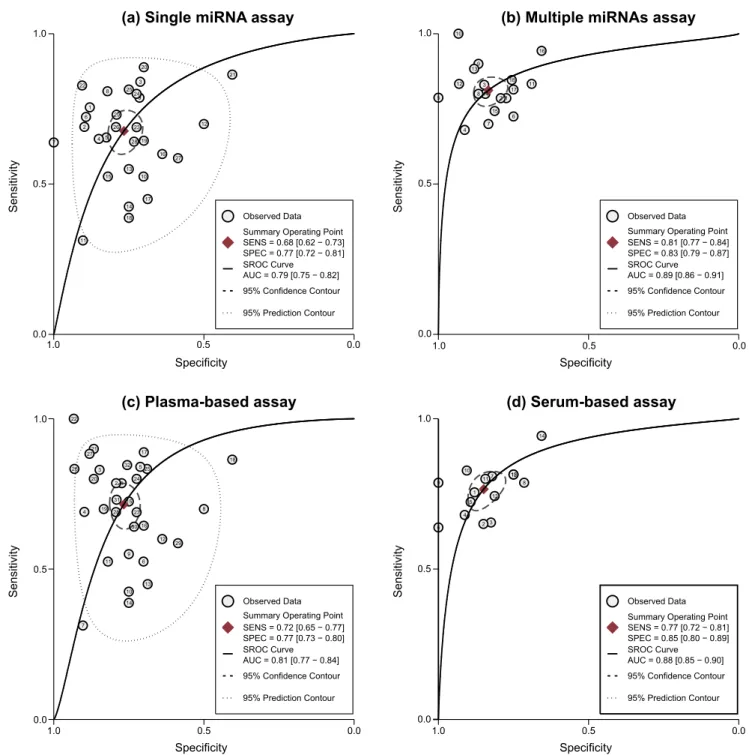
Figure 4. SROC curve with pooled estimates of sensitivity, specificity and AUC on the diagnostic value of circulating miRNAs in GC detection (a: single-miRNA assay; b: multiple-miRNAs assay; c: plasma-based assay; d: serum-based assay).
doi:10.1371/journal.pone.0113401.g004
Circulating MiRNAs for Gastrointestinal Tumors
As for GC, since significant heterogeneity between studies was observed in sensitivity and specificity data (I2= 84.53% and I2= 78.98%, respectively), the random-effects model was used. The pooled parameters calculated from all 47 studies on GC were as follows: SEN, 0.77 (95% CI: 0.72–0.80); SPE, 0.81 (95% CI: 0.77–0.84); PLR, 4.0 (95% CI: 3.2–4.9); NLR, 0.29 (95% CI: 0.24–0.36); and DOR, 14 (95% CI: 9–20). Figure 3B shows the corresponding SROC curve with the AUC of 0.86 (95%CI: 0.82– 0.88), indicating that circulating miRNAs may be able to differentiate GC patients from controls with a relatively high accuracy. Subgroup analysis based on different type of miRNA
assay suggested that multiple-miRNAs assay showed superior diagnostic properties (Figure 4B) than single one (Figure 4A), with SEN of 0.87 versus 0.75, SPE of 0.84 versus 0.80, and AUC of 0.92 versus 0.84 (Table 2). Notably, we found that plasma-based assay (Figure 4C) has a higher accuracy compared with serum-based assay (Figure 4D), suggesting that plasma is a better matrix for miRNA detection.
Similarly, the random-effects model was used for meta-analysis of studies on CRC since significant heterogeneity existed (I2= 87.18% andI2= 83.27%, respectively). The pooled estimates all 47 studies on CRC are as follows: SEN of 0.73 (95% CI: 0.69–
Figure 5. SROC curve with pooled estimates of sensitivity, specificity and AUC on the diagnostic value of circulating miRNAs in CRC detection (a: single-miRNA assay; b: multiple-miRNAs assay; c: plasma-based assay; d: serum-based assay).
doi:10.1371/journal.pone.0113401.g005
Circulating MiRNAs for Gastrointestinal Tumors
0.77), SPE of 0.80 (95% CI: 0.76–0.83), PLR of 3.6 (95% CI: 3.0– 4.2), NLR of 0.34 (95% CI: 0.29–0.39), and DOR of 11 (95% CI: 8–14) (Table 2). Figure 3C shows the corresponding SROC curve with the AUC of 0.83 (95%CI: 0.80–0.86), indicating that the diagnostic accuracy of miRNAs in CRC detection is slightly worse
than the overall diagnostic performance of miRNAs in gastro-intestinal cancers detection. Similar to miRNAs in GC detection, we found that multiple-miRNAs assay (Figure 5B) in differentiat-ing CRC patients from controls achieve a better diagnostic performance than single-miRNA assay (Figure 5A). However,
Figure 6. Forest plots of multivariable meta-regression analyses for sensitivity and specificity (a: single-miRNA assay; b: multiple-miRNAs assay).
doi:10.1371/journal.pone.0113401.g006
Circulating MiRNAs for Gastrointestinal Tumors
contrast to the results of GC, serum-based miRNAs assay (Figure 5D) has a higher accuracy compared with plasma-based assay (Figure 5C) in CRC detection.
Meta-regression and publication bias
The meta-regression analyses for both GC (Figure 6A) and CRC (Figure 6B) were performed to analyze the potential sources of inter-study heterogeneity. Overall, miRNA profiling and sample types may be the major sources of heterogeneity for miRNAs assay either in GC or CRC detection. To assess publication bias of included studies, the Deeks’ funnel plot asymmetry test was conducted (Figure 7). As shown in Figure 7, the slope coefficient was associated with aP-value of 0.38 for GC, 0.41 for CRC, 0.78 for EC, suggesting a low likelihood of publication bias in our meta-analysis.
Discussion
Gastrointestinal cancers, including esophageal, gastric, and colorectal cancer, together with breast and lung tumors, are responsible for the most cancer-related mortality [1]. Although endoscopic examinations and random biopsies are currently the most reliable screening tool for gastrointestinal cancers, their invasive, unpleasant, and inconvenient nature as well as potential sampling errors have hampered the wide clinic application. Conventional cancer-specific biomarkers are sufficiently simple and fast; unfortunately, their diagnostic performances have mostly been insufficient for application as a primary tool in population-based screening. Therefore, large number of studies on the search for ideal candidate biomarkers of tumors is still ongoing. During the past few years, miRNAs have become the focus of most recent efforts in cancer research. Since their discovery, emerging evidences suggest that the miRNAs may play important role in tumor suppressing, since the aberrant expression of miRNA was discovered between cancer patients and healthy controls [73,74]. Subsequently, miRNAs as molecular markers have attracted much attention in cancer diagnosis [20,21,44,54,67].
Circulating miRNAs, also known as cell-free miRNAs, have a promising future as a novel class of reliable minimally invasive biomarkers for early cancer diagnosis due to their remarkable stability, relatively easy detection, and convenience to measure its sensitivity and specificity [73]. Since circulating miRNAs in serum
was first reported [75], a tremendous growth of interest is attracted to the feasibility of circulating miRNAs as potential biomarkers in gastrointestinal malignancies diagnosis. However, inconsistencies are still existed in the literature reviews regarding the reliability of circulating miRNAs for early detection of gastrointestinal cancers. The variations and discrepancies in individual studies are possibly due to their small sample sizes, variations in miRNA assay, and different type of cancers. In this meta-analysis, we summarize the recent findings focusing on the potential of circulating miRNAs as diagnostic biomarkers in gastrointestinal tumors, including esophageal, gastric, and colorectal cancer.
To the best of our knowledge, this is the first evidence-based meta-analysis to evaluate the diagnostic value of circulating miRNA on gastrointestinal malignancies. The pooled results based on all included studies showed circulating miRNAs yielding an AUC of 0.85 with 75% sensitivity and 81% specificity in discriminating gastrointestinal cancer patients from controls. Although the origin and function of circulating miRNAs in cancer diagnosis have not been systematically elucidated, they have displayed a superior diagnostic performance compared with conventional blood biomarkers like CEA (AUC of 0.549) for EC and CA19-9 for GC (AUC of 0.60). Accordingly, further studies are required to elucidate the mechanism and target of miRNAs and their roles in cellular and molecular pathways. Interestingly, our results suggest that plasma-based miRNA assay reaches a higher accuracy than serum-based one for GC; the conclusion is, however, opposite for CRC. The origin of source-related difference is still unclear and might be explained by unknown mechanism. Thus, large scale investigations are needed in the following study to determine whether the source-related differ-ences truly exist.
Another interesting finding of our study is that single-miRNA assay displayed a relatively low diagnostic performance with the AUC values of 0.84 for gastric cancer (GC) and 0.79 for colorectal cancer (CRC), while multiple-miRNAs assay significantly im-proved the diagnosing accuracy with AUC rising to 0.92 for GC and 0.89 for CRC, implying that the advantage of using combination of miRNAs to obtain a complete picture. It is widely accepted that using single tumor-related miRNA as disease fingerprints is much simpler and more straightforward than comprehensively detecting panels of miRNAs, but the specificity of biomarkers based on single miRNA is relatively poor. The
Figure 7. The Deeks’ test of the diagnostic meta-analysis (a: Deeks’ funnel for GC; b: Deeks’ funnel for CRC; c: Deeks’ funnel for EC.
The dotted line indicates the regression line. No publication bias was detected for this meta-analysis (GC:P= 0.38; CRC:P= 0.41; EC:P= 0.78). doi:10.1371/journal.pone.0113401.g007
Circulating MiRNAs for Gastrointestinal Tumors
molecular basis for the limitation of single miRNA as a tumor biomarker is that aberrant levels of single miRNA might be associated with several different types of cancers [76]. Further-more, cancer develops can be regarded as a result of complex multi-stage process of epigenetic and genomic abnormalities, and thus, should be targeted by multiple miRNAs [77]. Accordingly, employing panels of miRNAs instead of an individual miRNA as biomarkers represents a rational option to circumvent the limitations in utilizing miRNAs as a non-invasive blood-based biomarker in cancer detection, especially for the localized pathological conditions, where regular biopsies are hard to get.
Although circulating miRNAs have a promising potential as relevant novel non-invasive cancer biomarkers in future as shown in the current study, several limitations need to be addressed. First, methodologies for an accurate absolute quantification of miRNAs suffer from a lack of convention, which limits the cross-comparison between studies performed by different laboratories. Standardized protocol, which should be preferably followed across all studies, needs to be established aiming to minimize protocol-based bias. In addition, some researchers have showed correlations of grade and stage of cancers with specific circulating miRNAs. Therefore, further studies addressing the relationships between miRNAs expression and clinical/pathological parameters are very impor-tant and desirable. Third, most included studies in this meta-analysis only distinguished the cancer patients from healthy controls. It is vital to identify and develop panels of miRNAs that
can distinguish cancer from other diseases, especially from those with similar symptom diseases. Last but not least, as shown in Table 1, most of included studies were on Asian and little bit on Caucasian/African populations. Therefore, further studies on Caucasian/African populations may be needed.
Based on recent observations in gastrointestinal cancer, we conclude that circulating miRNAs, particularly the combination of multiple-miRNAs, may present as promising minimally invasive approach for the diagnosis and monitoring of gastrointestinal tumors. Further large-scale prospective studies are necessary to validate their potential applicability in human cancer diagnosis.
Supporting Information
Supplement S1 PRISMA Checklist. (DOC)
Supplement S2 QUADAS-2 Checklist. (PDF)
Author Contributions
Conceived and designed the experiments: AX. Performed the experiments: RW HW. Analyzed the data: YCX QLC. Contributed reagents/ materials/analysis tools: Yi Luo Yiqin Lin Yu Luo. Wrote the paper: RW HW YCX.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, et al. (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90.
2. Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11–30.
3. Suzuki H, Gotoda T, Sasako M, Saito D (2006) Detection of early gastric cancer: misunderstanding the role of mass screening. Gastric Cancer 9: 315–319. 4. McFarland EG, Levin B, Lieberman DA, Pickhardt PJ, Johnson CD, et al.
(2008) Revised colorectal screening guidelines: joint effort of the American Cancer Society, U.S. Multisociety Task Force on Colorectal Cancer, and American College of Radiology. Radiology 248: 717–720.
5. Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ (2009) Gastric cancer. Lancet 374: 477–490.
6. Takahashi S, Hirayama M, Kuroiwa G, Kawano Y, Takada K, et al. (2013) Diagnostic validity of CT gastrography versus gastroscopy for primary lesions in gastric cancer: evaluating the response to chemotherapy, a retrospective analysis. Gastric Cancer 16: 543–548.
7. Brenner H, Chang-Claude J, Jansen L, Knebel P, Stock C, et al. (2014) Reduced risk of colorectal cancer up to 10 years after screening, surveillance, or diagnostic colonoscopy. Gastroenterology 146: 709–717.
8. Neri E, Faggioni L, Cerri F, Turini F, Angeli S, et al. (2010) CT colonography versus double-contrast barium enema for screening of colorectal cancer: comparison of radiation burden. Abdom Imaging 35: 596–601.
9. Duffy MJ (2007) Role of tumor markers in patients with solid cancers: A critical review. Eur J Intern Med 18: 175–184.
10. Choong MK, Tsafnat G (2012) Genetic and epigenetic biomarkers of colorectal cancer. Clin Gastroenterol Hepatol 10: 9–15.
11. Tarro G, Perna A, Esposito C (2005) Early diagnosis of lung cancer by detection of tumor liberated protein. J Cell Physiol 203: 1–5.
12. Sorio C, Mauri P, Pederzoli P, Scarpa A (2006) Non-invasive cancer detection: strategies for the identification of novel cancer markers. IUBMB Life 58: 193– 198.
13. Dbouk HA, Tawil A, Nasr F, Kandakarjian L, Abou-Merhi R (2007) Significance of CEA and VEGF as Diagnostic Markers of Colorectal Cancer in Lebanese Patients. Open Clin Cancer J 1: 1–5.
14. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854.
15. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297.
16. Davis BN, Hata A (2009) Regulation of MicroRNA Biogenesis: A miRiad of mechanisms. Cell Commun Signal 7: 18.
17. Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, et al. (2010) The microRNA spectrum in 12 body fluids. Clin Chem 56: 1733–1741. 18. Chen X, Ba Y, Ma L, Cai X, Yin Y, et al. (2008) Characterization of
microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006.
19. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, et al. (2002) Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99: 15524– 15529.
20. Ng EK, Chong WW, Jin H, Lam EK, Shin VY, et al. (2009) Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut 58: 1375–1381. 21. Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, et al. (2010)
Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer 102: 1174–1179.
22. Zhang C, Wang C, Chen X, Yang C, Li K, et al. (2010) Expression profile of microRNAs in serum: a fingerprint for esophageal squamous cell carcinoma. Clin Chem 56: 1871–1879.
23. Heneghan HM, Miller N, Kelly R, Newell J, Kerin MJ (2010) Systemic miRNA-195 differentiates breast cancer from other malignancies and is a potential biomarker for detecting noninvasive and early stage disease. Oncologist 15: 673– 682.
24. Keller A, Leidinger P, Borries A, Wendschlag A, Wucherpfennig F, et al. (2009) miRNAs in lung cancer - studying complex fingerprints in patient’s blood cells by microarray experiments. BMC Cancer 9: 353.
25. Li LM, Hu ZB, Zhou ZX, Chen X, Liu FY, et al. (2010) Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res 70: 9798–9807.
26. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105: 10513–10518.
27. Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M (2008) MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg 12: 2171–2176.
28. Leeflang MM, Deeks JJ, Gatsonis C, Bossuyt PM (2008) Systematic reviews of diagnostic test accuracy. Ann Intern Med 149: 889–897.
29. Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, et al. (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155: 529–536.
30. Deeks JJ (2001) Systematic reviews in health care: Systematic reviews of evaluations of diagnostic and screening tests. BMJ 323: 157–162.
31. Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM (2003) The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 56: 1129– 1135.
32. Higgins JP, Thompson SG, Deeks JJ, Altman DG (2003) Measuring inconsistency in meta-analyses. BMJ 327: 557–560.
33. Jackson D, White IR, Thompson SG (2010) Extending DerSimonian and Laird’s methodology to perform multivariate random effects meta-analyses. Stat Med 29: 1282–1297.
Circulating MiRNAs for Gastrointestinal Tumors
34. Deeks JJ, Macaskill P, Irwig L (2005) The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58: 882–893.
35. Zhou H, Guo JM, Lou YR, Zhang XJ, Zhong FD, et al. (2010) Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using microRNA as a marker. J Mol Med (Berl) 88: 709–717.
36. Liu R, Zhang C, Hu Z, Li G, Wang C, et al. (2011) A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 47: 784–791.
37. Zheng Y, Cui L, Sun W, Zhou H, Yuan X, et al. (2011) MicroRNA-21 is a new marker of circulating tumor cells in gastric cancer patients. Cancer Biomark 10: 71–77.
38. Zhu CJ, Zhang JY, Wei J, Liu YQ, Shu YQ (2011) [MiR-191 and miR-29a in plasma are promising novel biomakers for detection of gastric cancer]. Acta Universitatis Medicinalis Nanjing(Natural Science) 31: 1173–1178.
39. Konishi H, Ichikawa D, Komatsu S, Shiozaki A, Tsujiura M, et al. (2012) Detection of gastric cancer-associated microRNAs on microRNA microarray comparing pre- and post-operative plasma. Br J Cancer 106: 740–747. 40. Li BS, Zhao YL, Guo G, Li W, Zhu ED, et al. (2012) Plasma microRNAs,
miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One 7: e41629.
41. Li ZQ, Huang PW, Zhu CJ, Chen CP, Cheng T, et al. (2012) [Diagnostic value of expressions of miR-27a and miR-181b for early detection of gastric cancer]. Jiangsu Med J 38: 1665–1667.
42. Liu H, Zhu L, Liu B, Yang L, Meng X, et al. (2012) Genome-wide microRNA profiles identify miR-378 as a serum biomarker for early detection of gastric cancer. Cancer Lett 316: 196–203.
43. Song MY, Pan KF, Su HJ, Zhang L, Ma JL, et al. (2012) Identification of serum microRNAs as novel non-invasive biomarkers for early detection of gastric cancer. PLoS One 7: e33608.
44. Valladares-Ayerbes M, Reboredo M, Medina-Villaamil V, Iglesias-Diaz P, Lorenzo-Patino MJ, et al. (2012) Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J Transl Med 10: 186.
45. Wang B, Zhang Q (2012) The expression and clinical significance of circulating microRNA-21 in serum of five solid tumors. J Cancer Res Clin Oncol 138: 1659–1666.
46. Zhou H, Xiao B, Zhou F, Deng H, Zhang X, et al. (2012) MiR-421 is a functional marker of circulating tumor cells in gastric cancer patients. Biomarkers 17: 104–110.
47. Cai H, Yuan Y, Hao YF, Guo TK, Wei X, et al. (2013) Plasma microRNAs serve as novel potential biomarkers for early detection of gastric cancer. Med Oncol 30: 452.
48. Li C, Li JF, Cai Q, Qiu QQ, Yan M, et al. (2013) miRNA-199a-3p in plasma as a potential diagnostic biomarker for gastric cancer. Ann Surg Oncol 20 Suppl 3: S397–405.
49. Sheinerman KS, Tsivinsky VG, Umansky SR (2013) Analysis of organ-enriched microRNAs in plasma as an approach to development of Universal Screening Test: feasibility study. J Transl Med 11: 304.
50. Shiotani A, Murao T, Kimura Y, Matsumoto H, Kamada T, et al. (2013) Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br J Cancer 109: 2323–2330.
51. Xu Q, Dong QG, Sun LP, He CY, Yuan Y (2013) Expression of serum miR-20a-5p, let-7a, and miR-320a and their correlations with pepsinogen in atrophic gastritis and gastric cancer: a case-control study. BMC Clin Pathol 13: 11. 52. Chen Q, Ge X, Zhang Y, Xia H, Yuan D, et al. (2014) Plasma miR-122 and
miR-192 as potential novel biomarkers for the early detection of distant metastasis of gastric cancer. Oncol Rep 31: 1863–1870.
53. Peng WZ, Ma R, Wang F, Yu J, Liu ZB (2014) Role of miR-191/425 Cluster in Tumorigenesis and Diagnosis of Gastric Cancer. Int J Mol Sci 15: 4031–4048. 54. Zhu C, Ren C, Han J, Ding Y, Du J, et al. (2014) A five-microRNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer.
55. Huang Z, Huang D, Ni S, Peng Z, Sheng W, et al. (2010) Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. International Journal of Cancer 127: 118–126.
56. Pu XX, Huang GL, Guo HQ, Guo CC, Li H, et al. (2010) Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 25: 1674–1680.
57. Liu HS, Ma YY, Xiao HS (2012) [The diagnostic value of serum microRNAs including miR-129-3p, miR-767-3p and miR-877* for colorectal cancer]. Tumor 32: 42–48.
58. Wang Q, Huang Z, Ni S, Xiao X, Xu Q, et al. (2012) Plasma miR-601 and miR-760 are novel biomarkers for the early detection of colorectal cancer. PLoS ONE 7: e44398.
59. Feng L, Pang Z, Sha S, Wu B, Xu F, et al. (2013) [The value of diagnosis and prognosis prediction of serum miR-29a and miR-92a for colorectal cancer]. Chin J Clin Oncol Rehabil: 1313–1315.
60. Giraldez MD, Lozano JJ, Ramirez G, Hijona E, Bujanda L, et al. (2013) Circulating MicroRNAs as biomarkers of colorectal cancer: Results from a genome-wide profiling and validation study. Clinical Gastroenterology and Hepatology 11: 681–688.e683.
61. Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, et al. (2013) Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol 34: 2175–2181.
62. Luo X, Stock C, Burwinkel B, Brenner H (2013) Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS ONE 8: e62880.
63. Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, et al. (2013) Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. Journal of the National Cancer Institute 105: 849–859.
64. Wang S, Xiang J, Li Z, Lu S, Hu J, et al. (2013) A plasma microRNA panel for early detection of colorectal cancer. Int J Cancer.
65. Yong FL, Law CW, Wang CW (2013) Potentiality of a triple microRNA classifier: MiR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 13.
66. Zhang GJ, Zhou T, Liu ZL, Tian HP, Xia SS (2013) Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Molecular and Clinical Oncology 1: 379–384.
67. Zanutto S, Pizzamiglio S, Ghilotti M, Bertan C, Ravagnani F, et al. (2014) Circulating miR-378 in plasma: a reliable, haemolysis-independent biomarker for colorectal cancer. Br J Cancer 110: 1001–1007.
68. Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, et al. (2011) Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 105: 104–111.
69. Zhang T, Wang Q, Zhao D, Cui Y, Cao B, et al. (2011) The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci (Lond) 121: 437–447.
70. Hirajima S, Komatsu S, Ichikawa D, Takeshita H, Konishi H, et al. (2013) Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br J Cancer 108: 1822–1829.
71. Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, et al. (2013) Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer 108: 644– 652.
72. Zhang T, Zhao D, Wang Q, Yu X, Cui Y, et al. (2013) MicroRNA-1322 regulates ECRG2 allele specifically and acts as a potential biomarker in patients with esophageal squamous cell carcinoma. Mol Carcinog 52: 581–590. 73. Redova M, Sana J, Slaby O (2013) Circulating miRNAs as new blood-based
biomarkers for solid cancers. Future Oncol 9: 387–402.
74. Shen J, Stass SA, Jiang F (2013) MicroRNAs as potential biomarkers in human solid tumors. Cancer Lett 329: 125–136.
75. Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, et al. (2008) Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141: 672–675. 76. Sita-Lumsden A, Dart DA, Waxman J, Bevan CL (2013) Circulating
microRNAs as potential new biomarkers for prostate cancer. Br J Cancer 108: 1925–1930.
77. Zen K, Zhang CY (2012) Circulating MicroRNAs: A novel class of biomarkers to diagnose and monitor human cancers. Medicinal Research Reviews 32: 326– 348.
Circulating MiRNAs for Gastrointestinal Tumors