w ww.e l s e v i e r . c o m / l o c a t e / b j p
Original
Article
Identification
of
reference
genes
for
gene
expression
normalization
in
safflower
(
Carthamus
tinctorius
)
Fei
Liu
a,
Dan
Dan
Guo
a,
Yan
Hua
Tu
a,
Ying
Ru
Xue
a,
Yue
Gao
b,∗,
Mei
Li
Guo
a,∗aDepartmentofPharmacognosy,CollegeofPharmacy,SecondMilitaryMedicalUniversity,Shanghai,China
bTeachingExperimentalCenter,CollegeofPharmacy,SecondMilitaryMedicalUniversity,Shanghai,China
a
r
t
i
c
l
e
i
n
f
o
Articlehistory:
Received5June2015
Accepted24May2016
Availableonline16June2016
Keywords:
Safflower
Referencegene
RT-qPCR
a
b
s
t
r
a
c
t
Safflower(CarthamustinctoriusL.,Asteraceae)isanimportantoilcropandmedicinalplant.Gene expres-sionanalysisisgainingimportanceintheresearchofsafflower.QuantitativePCRhasbecomeapowerful methodforgenestudy.ReferencegenesareoneofthemajorqualificationrequirementsofqPCRbecause theycanreducethevariability.Toidentifythereferencegenesinsafflower,ninecandidategenesof thehousekeepinggeneswereselectedfromtheESTlibraryofsafflowerconstructedbyourlab:CtACT (actin),CtGAPDH(glyceraldehyde3-phosphatedehydrogenase),CtE1F4A(elongationfactor1alpha), CtTUA(alpha-tubulin),CtTUB(beta-tubulin),CtPP2A (serine/threonine-proteinphosphatase),CtE1F4A (eukaryoticinitiationfactor4A),CtUBI(Ubiquitin),andCt60S(60Sacidicribosomalprotein). Expres-sionstabilitywasexaminedbyqPCRacross54samples,representingtissuesatdifferentfloweringstages andtwochemotypeofsafflowerlines.Weassessedtheexpressionstabilityofthesecandidategenesby employingfourdifferentalgorithms(geNorm,NormFinder,Ctapproach,andBestKeeper)andfound thatCtUBIandCt60Swerethehighlyrankedcandidategenes.CtUBIandCt60Swereusedasreference genestoevaluatetheexpressionofCtFAD2-10andCtKASII.OurdatasuggestCtUBIandCt60Scouldbe usedasinternalcontrolstonormalizegeneexpressioninsafflower.
©2016SociedadeBrasileiradeFarmacognosia.PublishedbyElsevierEditoraLtda.Thisisanopen accessarticleundertheCCBY-NC-NDlicense(http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Safflower(CarthamustinctoriusL.,Asteraceae)isathistle-like, self-compatible, annual, diploid (2n=24) herbaceous crop that thrives in hot, dry climates and survives on minimal surface moisture.Thesafflowercultivarsaredistributedfromthe Mediter-raneantothePacificOceanatlatitudesbetween20◦Sand40◦N,
whereverahot,dryclimatesuitsthecrop.Insomecountries, saf-flowerhasbecomeanimportantcropduetotherichcontentof edibleoil,whichhasthehighestpolyunsaturated/saturatedratios ofanyoilavailable(Gecgeletal.,2007;Yeilaghietal.,2012). Saf-flowerisalsoavaluablemedicinalplant.Theflowersofsafflower canbeusedforthetreatmentofcardiovascularand cerebrovascu-lardiseases(Tianetal.,2010;AsgarpanahandKazemivash,2013) andtheextractsfromsafflowercanbeusedasanti-inflammatory agentsaswell(Junetal.,2011).Thereareseveralstudiesabout thegeneticvariationofsafflowercultivarsusingvariousmolecular DNAmarkers,suchasSNP(ChapmanandBurke,2007),SRAP(Peng etal.,2008),ISSR(Chapmanetal.,2009;Golkaretal.,2011),and
∗ Correspondingauthors.
E-mails:13701638627@163.com(Y.Gao),mlguo@126.com(M.L.Guo).
AFLP(Zhangetal.,2009;Fengetal.,2010;Lietal.,2010). Mean-while,inthepreviousstudyofourlab,wehavefoundthatobvious differentiationhasoccurredinsafflowerpopulationsfromexterior appearancetoinnerchemicalconstituent,duetothelong natu-ralandartificialselection(Pengetal.,2008;Zhangetal.,2009; Fengetal.,2010;Lietal.,2010).Fromourconclusion, hydroxysaf-floryellowA(HYSA),astheactivecompound,isthemainfactorto determinethediversityofsafflower(Yangetal.,2011).
Assecondarymetabolitesinsafflower,theflavonoidsaremajor componentsoftheextractsfromtheflowerswithmedicinal func-tion(AndersenandMarkham,2010;AsgarpanahandKazemivash, 2013).HSYA,oneofthemostimportantflavonoidswithaunique presenceintheflowerpetalsofsafflower,playsamajorrolein thepharmacologicaleffectsofflavonoids(Fengetal.,2013;Sun et al., 2013; Wang et al., 2013). The flowering process of saf-flowerisacomplexdevelopmentassociatedwiththebiosynthesis offlavonoids,particularlythecolorchange(Tanakaetal.,2010). Insafflowerseeds,theidentificationandinitialcharacterizationof theFAD2genefamilywithelevenmembersprovidesaninsight intotheprincipaldeterminantsofsynthesisoflinoleicacidin saf-flowerseedoil(Caoetal.,2013;Liuetal.,2013).AndtheCtFAD3
enzymeactivityisimportantforfattyaciddesaturationinsafflower flower(Guanetal.,2014).Theunderstandingoftheexpressionof
http://dx.doi.org/10.1016/j.bjp.2016.05.006
0102-695X/©2016SociedadeBrasileiradeFarmacognosia.PublishedbyElsevierEditoraLtda.ThisisanopenaccessarticleundertheCCBY-NC-NDlicense(http://
thekeygeneswillhelpinvestigatethemechanisminvolvedinthe biosynthesisofflavonoidsandfattyacids.
Quantitativereal-timePCR(qPCR)isaroutinetoolwithhigh sensitivityand specificityfor thequantificationofgene expres-sion(Gachonetal.,2004).Whenwestudythegeneexpression, thereferencegenescanbeusedinthenormalizationof experi-mentalvariations,suchastheamountofmaterial,extractionof RNA,efficiencyofreversetranscription, andsoon(Silver etal., 2006;Gutierrezetal.,2008;Bustinetal.,2009;Guéninetal.,2009). Itisimportanttonormalizetheexpressionofthetargetgenein ordertoobtainreliableand accurateresultsbyusingthe refer-encegenes. Housekeeping genes areoften considered asbeing constantlyexpressedandarealwaysusedasinternalstandards. Butmanyresearchesshowedthattheexpressionofhousekeeping genesvariesindifferentmaterialsandeveninthesame materi-alsduringdifferenttreatments(Thellinetal.,1999;Schmidtand Delaney,2010).Sotheevaluationofreferencegenesindifferent planttissuesduringbioticorabioticstressisnecessaryfor expres-sionstudies.
Housekeepinggenes,suchasactin(Basetal.,2004),elongation factor1alpha(SchmidtandDelaney,2010;Lietal.,2012), alpha-tubulin(JarosovaandKundu,2010;Wanetal.,2011),beta-tubulin (JarosovaandKundu,2010),serine/threonine-proteinphosphatase (Liu et al., 2012), eukaryotic initiationfactor 4A (Silveiraet al., 2009),ubiquitin(Infanteetal.,2008), and60Sacidic ribosomal protein(Leetal.,2012),arecommonly usedasreferencegenes forgene expressionstudies.Therearestudieswhich haveused referencegenesforthenormalizationofgeneexpressionin saf-flower(Lietal.,2012;Caoetal.,2013).Butthesereferencegenes were not evaluated for qPCR data in safflower tissues. In this study,nine housekeepinggenesnamelyCtACT(actin), CtGAPDH
(glyceraldehyde 3-phosphate dehydrogenase), CtE1F4A (elonga-tionfactor1alpha),CtTUA(alpha-tubulin),CtTUB(beta-tubulin),
CtPP2A(serine/threonine-proteinphosphatase),CtE1F4A (eukary-oticinitiationfactor4A),CtUBI(Ubiquitin),andCt60S(60Sacidic ribosomal protein) were selected as candidate reference genes fromourconstructededtranscriptome dataof safflower.Atthe sametime,tworepresentativechemotypeofsafflowervarieties, with-HSYA(yellowflower)andwithout-HSYA(whiteflower)were selectedtoidentifyreferencegenes.Theexpressionofthesegenes in the bract, ovary, stem, leaf, calyx,petal (I–IV, fourdifferent stages ofdevelopmentduring flowering)of twosafflower lines wereanalyzed using four differentalgorithms, that is, geNorm (Vandesompeleetal.,2002),NormFinder(Andersenetal.,2004),
Ctapproach (Silveret al.,2006), andBestKeeper (Pfaffl etal., 2004).Ourstudywillhelptoachievemoreaccurateandreliable resultsinawidevarietyofsafflowersamples.
Materialsandmethods
Plantmaterial
Theseedsoftwolinesofsafflower,CarthamustinctoriusL., Aster-aceae(ZHH0082withyellowflowerandXinHonghuaNO.7with whiteflower)fromtheChinesepopulationswerecultivatedinthe fieldatthemedicinalbotanicalgardenofSecondMilitary Medi-calUniversity.Thesamples(bract,ovary,stem,leaf,calyx,petal (I–IV))werecollectedfromthreedifferentfloweringplants (bio-logicaltriplicates)andimmediatelyfrozeninliquidnitrogen.All 54sampleswerethenstoredat−70◦CforRNAextraction.
TotalRNAextraction
Total RNA was isolated using the RNA Extraction Plant Mini Kit (LifeFeng, Shanghai, China) according to the protocol
providedbythemanufacturer.RNAconcentrationandqualitywere measuredwiththeNanoDropspectrophotometer(NanoDrop Tech-nologies)andagarosegelelectrophoresis.OnlytheRNAsamples withA260/A280 ratiosbetween1.9 and 2.1 and A260/A230 ratios
greaterthan2.0wereusedforcDNAsynthesis.
FirststrandcDNAssynthesis
FirststrandcDNAsweresynthesizedaccordingtothe manufac-turer’sinstructionsofTransScript®One-StepgDNARemovaland
cDNASynthesisSuperMix(TransGenBiotech,Beijing,China).Total RNA(1g),suitablevolumesofH2O,and1lanchoredOligo(dT)
18primer(0.5g/l)weremixedandincubatedat65◦Cfor5min
followedbycoolingonice.Thereversetranscriptasereactionswere startedafteradding10l2×TSreactionmix,1lEnzymeMix,and
1lgDNARemoverat42◦Cfor45min.Andthenthemixturewas
heatedfor5minat85◦Cforinactivatingtheenzymes.AllcDNA
sampleswerediluted1:10withRNase-freewaterbeforebeingused astemplatesintheqPCRanalysis.
Q-PCR
Ninesequenceswereselectedascandidatereferencegenesfrom thepetalESTlibrariesofC.tinctorius(Table1).TheqPCRprimers weredesignedusingtheBeaconDesignerv8.0software.Amplicon lengthsvariedfrom75to200bp,withmeltingtemperatures(Tm) varyingbetween52◦Cand60◦Candprimerlengthsbetween18
and22bp.PrimerpairsweretestedforspecificitybyqPCR,followed byadissociationcurveandagarosegelelectrophoresis(Fig.S1). PCRreactionswereperformedin96-wellplateswiththeCFX96 TouchTMReal-TimePCRDetectionSystem(Bio-RadLaboratories,
CA,USA) andABI7500Real-Time PCR System(Applied Biosys-tems,FosterCity,USA).TheCFX96wasusedtoobtaintheqPCR dataofninecandidategenesandABI7500wasusedtoobtainthe qPCRdataofCtFAD2-10(KC257456,F-CTTTACCGTATGGCTTTAG, R-GTGTGTTGAAGGTATGTG), CtKASII (KC257458, F-GACAGGTTTAT-GCTCTAC, R-CAATCAGAACTCCACATC), and two stable reference genes.Eachreactioncomprising20lofthefollowingwas pre-paredasfollows:0.3mforwardprimer,0.3mreverseprimer, 10l 2× TranStartTM Top Green qPCR SuperMix (TransGen
Biotech,Beijing, China),2lcDNA,0.4lpassivereference dye II(onlyused inABIsystem), and suitablevolumesof H2O.The
thermocyclingconditionsweresetat95◦C for30s,followedby
40cyclesof10sat95◦Cfortemplatedenaturation,15satTmfor
annealing,and20s(30sforABIsystem)at72◦Cforextensionand
fluorescencemeasurement.Afterwards,thedissociationcurvewas obtainedbyheatingtheampliconfrom60◦Cto95◦C and
read-ingateach0.5◦Cincrease(0.2◦CforABIsystem).Threetechnical
replicateswereperformedforeachPCRreaction.
Dataanalysis
Table1
NinecandidatereferencegenesandtheirprimersequencesforqPCR.
Geneidentification/genedescription EvalueID(%) Primersequence Ampliconsize Amplification efficiency
GeneBank Accession Number
Tm
CtACT 0.0 F-ACTGGTGTTATGGTAGGA 89 1.85 KJ634809 60
actin 100 R-GGATACTTCAAGGTAAGGATA
CtGAPDH 0.0 F-GTTGTGGACTTAACCGTAA 78 1.83 KJ634805 53
glyceraldehyde3-phosphatedehydrogenase 93 R-TTCTGATTCCTCCTTAATAGC
CtEF1 0.0 F-CCAAGAGACCATCAGACAA 76 1.88 KJ634806 58
elongationfactor1alpha 98 R-GGCACAGTTCCAATACCA
CtTUA 0.0 F-CTACACCAACCTCAATCG 91 1.89 KJ634803 58
␣-tubulin 99 R-AGTCACATCCACATTCAAG
CtTUB 0.0 F-GGAAGAGGAGTATGATGA 93 1.88 KJ634802 54
-tubulin 99 R-AATGGCAGTTGAGATTAC
CtPP2A 0.0 F-GAGAACCTGATGTAACGAGAC 80 1.89 KJ634804 58
serine/threonine-proteinphosphatase 95 R-ACCACCAAGCAAGCAATC
CtE1F4A 0.0 F-CGCTGATTACATTAAGATG 90 1.85 KJ634807 54
eukaryoticinitiationfactor4A 98 R-AATTGGAAGATATCGTAGAT
CtUBI 3e−87 F-TCACTTATGTTTACCAGAA 150 1.85 KJ634808 54
Ubiquitin 98 R-GCTTTCAATTTCAACTCA
Ct60S 9e−165 F-CATCCATTATCCAACAATC 92 1.86 KJ634810 54
60Sacidicribosomalprotein 88 R-AAGAGTAATCAGTCTCCA
Results
SelectionofputativereferencegenesforqPCRexperiments
Inordertofindthebestreferencegenesinsafflower,nine
house-keeping genes, CtACT, CtGAPDH, CtE1F4A, CtTUA, CtTUB, CtPP2A,
CtE1F4A,CtUBI,andCt60S,wereselectedasputativereferencegenes (Table 1).These genes were usedto BLAST search against a C. tinctoriusEST(expressedsequencetag)libraryconstructedbyour laboratoryfromthepetalduringflowering.Thehousekeepinggene commonlyusedasreferencegeneisthe18SrRNA.Inthisstudy, however,theCtvaluesof18Sinthesafflowertissueswereatleast 10cycleshigherthantheCtvaluesoftheothercandidategenes (datanot shown).Such highCtvalues make 18Sunsuitable for useasareferencegene(Lietal.,2012).Thedissociationcuresof ampliconsoftheseninecandidategenesexhibitedasinglepeak showedthattheprimerpairswerespecificity,whichwerealso ver-ifiedbyagarosegelelectrophoresisresults(Fig.S1).Theexpression ofthesegenesinbract,ovary,stem,leaf,calyx,petal(I–IV,four dif-ferentstagesofdevelopmentduringflowering,Fig.2,TableS1)was analyzed.Sincefloweringbeginsintheoutercircleoffloretsand progressescentripetallytowardthecenterofthecapitulum,the
fourstagesofpetalscanbefoundinonecapitulum.Thereasonwe differentiatethefourstagesofpetalsisthatthecompoundshave significantvariationduringthedevelopmentof theC. tinctorius
flowers(Salemetal.,2011)andthegeneexpressionsaredifferent aswell(Mallonaetal.,2010).Inourstudy,atotalof54samples(two linesofsafflower(ZHH0082andXinHonghuaNO.7),nineorgans, threebiologicalreplicates)wereusedtoevaluatethestabilityof putativereferencegenes.Thesignificantdifferencebetweenthe ZHH0082andXinHonghuaNO.7wasthecolorofflowers.The flow-ersofZHH0082wereyellowandtheflowersofXinHonghuaNO.7 werewhite.
Expressionstabilityofputativereferencegenesviadifferential statisticalanalyses
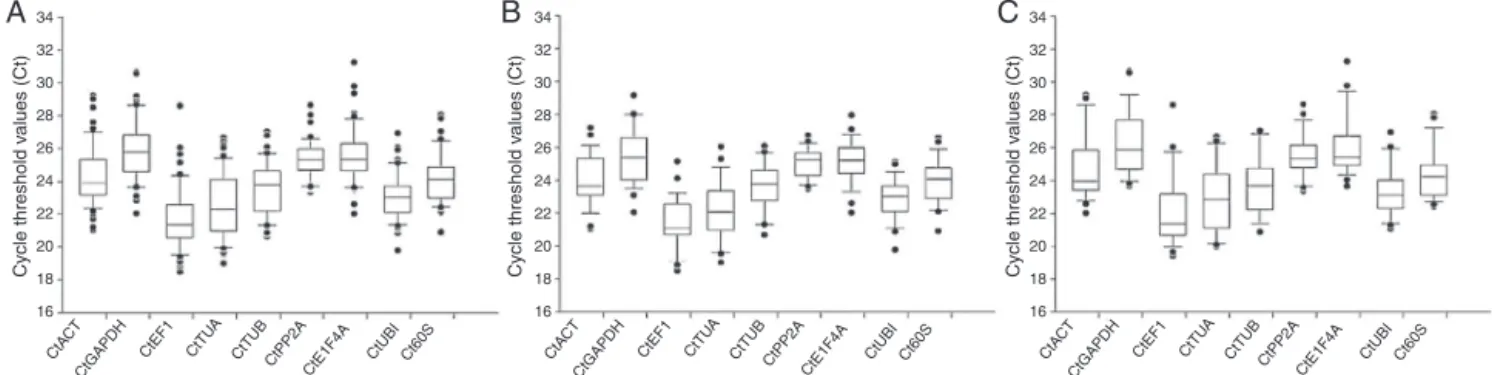
Thecyclethreshold(Ct)valuesprofilingofcandidategenesin differentorgansandsafflowerlinesareshowninFig.3.Themean ofeachoftheninecandidategenesin54samplesareshownin
TableS2.Inthisstudy,geNorm,NormFinder,Ctapproachand BestKeeperwereusedtoanalyzethestabilityofgeneexpression.
GeNormcalculatedthegeneexpressionstabilityvalueM(the average pairwise variation of a particular gene with all other
Table2
RankingofthecandidatereferencegenesaccordingtotheirvaluescalculatedbygeNorm,NormFinder,CtapproachandBestKeeperinAS,YandW.
Rank Allsamples(AS) Yellow(Y) White(W)
geNorm (M) NormFinder (SV)
Ct
approach
BestKeeper geNorm (M) NormFinder (SV)
Ct
approach
BestKeeper geNorm (M) NormFinder (SV)
Ct
approach
BestKeeper
1 CtUBI Ct60S Ct60S CtPP2A Ct60S CtUBI Ct60s CtPP2A CtACT CtPP2A CtUBI CtPP2A
0.776 0.061 0.671 0.774 0.063 0.670 0.756 0.062 0.643
2 Ct60S CtACT CtACT CtUBI CtUBI Ct60s CtACT CtUBI CtTUB CtEF1 CtTUB CtUBI
0.777 0.066 0.673 0.781 0.070 0.672 0.759 0.072 0.649
3 CtACT CtUBI CtUBI Ct60S CtTUB CtACT CtTUB CtE1F4A CtGAPDH CtACT Ct60S Ct60S
0.788 0.068 0.681 0.788 0.081 0.683 0.767 0.092 0.651
4 CtTUB CtGAPDH CtTUB CtTUB CtACT CtE1F4A CtUBI Ct60S CtUBI Ct60S CtACT CtTUB
0.837 0.076 0.724 0.804 0.081 0.689 0.770 0.113 0.660
5 CtE1F4A CtEF1 CtE1F4A CtE1F4A CtEF1 CtTUB CtGAPDH CtTUB Ct60S CtGAPDH CtE1F4A CtE1F4A
0.839 0.078 0.736 0.814 0.085 0.748 0.773 0.123 0.699
6 CtGAPDH CtE1F4A CtGAPDH CtACT CtE1F4A CtGAPDH CtEF1 CtACT CtE1F4A CtTUA CtGAPDH CtGAPDH
0.845 0.085 0.748 0.815 0.088 0.772 0.780 0.137 0.700
7 CtEF1 CtTUA CtEF1 CtGAPDH CtGAPDH CtEF1 CtPP2A CtEF1 CtPP2A CtUBI CtPP2A CtACT
0.851 0.127 0.785 0.878 0.103 0.863 0.856 0.144 0.766
8 CtPP2A CtTUB CtPP2A CtTUA CtPP2A CtPP2A CtE1F4A CtGAPDH CtEF1 CtE1F4A CtEF1 CtTUA
0.975 0.127 0.818 1.021 0.148 0.869 0.880 0.146 0.781
9 CtTUA CtPP2A CtTUA CtEF1 CtTUA CtTUA CtTUA CtTUA CtTUA CtTUB CtTUA CtEF1
1.104 0.138 0.957 1.130 0.168 0.974 1.077 0.154 0.926
RankAggrega Ct60S/CtUBI/CtACT CtUBI/Ct60S/CtTUB CtUBI/Ct60S/CtTUB
7
A
B
3025
20
15
10
5
0 6
5
4
CtUBI+Ct60S CtUBI Ct60S 3
2
1
0
Relativ
e quantification of
CtF
AD2-10
Relativ
e quantification of
CtKASII
C
B G J L T I II III IV B C G J L T I II III IV
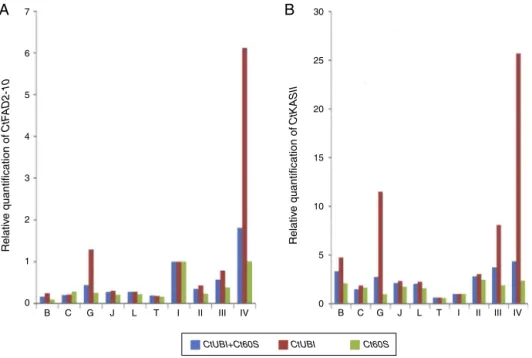
Fig.1. RelativequantificationofCtFAD2-10andCtKASIIexpressionusingdifferentinternalcontrolsanalyzedbythe2−Ctmethodinallsamples.Relativequantificationof
CtFAD2-10andCtKASIIexpressionweredetectedusingCtUBIandCt60Sbothindividuallyandincombination.TherelativeexpressionofpetalIwassetto1.(A)CtUBI,Ct60S
andthegeometricaverageofCtUBIandCt60SwereusedasinternalcontrolsforCtFAD2-10expression.(B)CtUBI,Ct60SandthegeometricaverageofCtUBIandCt60Swere
usedasinternalcontrolsforCtKASIIexpression.B,bract;C,calyx;L,leaf;O,ovary;S,stem;I–IV,petalI–IV.
A
B
C
3 cm
2 cm
1 cm
3 cm
2 cm
1 cm
I II III IV
I II III IV
F
D
E
Fig.2. Twolinesofsafflower.(1)A,B,CaretheZHH0082(Yellow);(2)D,E,FaretheXinHonghuaNO.7(White);(A,D)capitulumofsafflower;(B,E)longitudinalsectionof
capitulum;(C,F)fourdifferentstagesofthepetalduringflowering.Blackarrowspointtothesamplescollectedfromsafflower.B,bract;O,ovary;T,stem;L,leaf;C,calyx;P, petal.
controlgenes)for areferencegene. ThelowertheM value,the morestablyexpressedthegeneis.NormFinderusedamodel-based approachfor identifyingtheoptimalnormalizationgene(s).The intra-andintergroupvariationswerecalculatedandincludedin thegeneexpressionstabilityvalues. Geneswiththelowest val-ueshadthemoststableexpression.GeNormandNormFinderused the linear scale expression quantities, which can be calculated fromtheCtvaluesbyusingastandardcurveorcomparativeCT method(Schmittgenand Livak, 2008).Ctapproach compared theCtvariationofpairsofhousekeepinggeneswithinindividual
samplestoidentifythereferencegenes.ThegeneswithlowerCt
16
CtA CT
CtEF1 CtTU A
CtT UB
CtPP2A CtE1F4A
CtUBICt60S CtGAP
DH
18 20
Cycle threshold v
alues (Ct)
Cycle threshold v
alues (Ct)
Cycle threshold v
alues (Ct)
22 24 26 28 30 32 34
16
CtA CT
CtEF1 CtTU A
CtT UB
CtPP2A CtE1F4A
CtUBICt60S CtGAPDH
18 20 22 24 26 28 30 32 34
A
B
C
16
CtA CT
CtEF1 CtTU A
CtT UB
CtPP2A CtE1F4A
CtUBICt60S CtGAPDH
18 20 22 24 26 28 30 32 34
Fig.3. Ctvaluesofcandidatereferencegenes.ExpressiondatashowedtheCtvalues(blackspots)ofeachgene.Theverticalboxesrepresentthe25thand75thpercentiles
witherrorbars(whiskers).Thelinesacrosstheboxesaredepictedasthemedian.(A)Allsamplesofsafflower,(B)samplesofZHH0082(Yellow),and(C)samplesofXin
HonghuaNO.7(White).
InAS(all54samplesofthetwolinesofsafflower),CtUBI,Ct60S, andCtACTwerethethreemoststablyexpressedcandidategenes calculatedbygeNorm,NormFinder andCt.In thelistof Best-Keeper,thetopthreewereCtPP2A,CtUBI,andCt60S.Themoststably expressedgeneidentifiedbyBestKeeperwasCtPP2A,whichhad thehighestvariationintheresultsfromothermethods(geNorm ranked8,NormFinderranked9,andCtranked8).Thetopthree aggregateordercalculatedbyRankAggregwasCt60S/CtUBI/CtACT
inAS(Table2).
TheresultsofASwereanalyzedbysegregatingintotwo sub-groups, Y and W. The Y group included 27 samples collected fromZHH0082(Yellow)and Wcontained27samplesfromXin HonghuaNO.7(White).Thefouralgorithmsproducedsimilar rank-inglistsinASand Y.Butthere weremoredifferencesbetween two lines, Yand W.In Y,Ct60S was thefirst, second, and the first best-ranked gene in the geNorm, NormFinder, and Ct
approach,respectively.However,Ct60Swasrankedatfifth,fourth, and third in W. In the analysisof NormFinder, theposition of
CtPP2AhadremarkablevariationbetweenYandW,whichwasthe eighthinYbutthefirstinW.Althoughtherewerediscrepancies amongtheordersproducedbythesealgorithmsinthetwolines, theaggregateorders werethesame, namelyCtUBI/Ct60S/CtTUB
(Table2).
Theresultsanalyzedbyalgorithmsweredifferentinall Flow-ers(F),Yellow-Flower(YF,flowersofZHH0082),andWhite-Flower (WF,flowersof XinHonghua NO.7).Accordingtotheresultsof NormFinder,theCtE1F4Awasthesecondmoststablyexpressed geneinF,seventhinYF,andfirstinWF.Inparticular,CtE1F4Awas themoststablyexpressedgeneinYFcalculatedbythegeNorm, NormFinder,andCtapproach.TheupgradedpositionsofCtE1F4A
resultedinchangingoftheaggregateordersinSA,F,andYF.But theorderinWFwasCt60S/CtUBI/CtACT,whichwasthesameasthat inAS(Table3).
Ithasbeensuggested thattheuseoftwo ormorereference genesforRT-qPCR studiesmightgenerate morereliableresults (Vandesompeleet al.,2002).To determinetheoptimalnumber ofgenesrequiredforaccuratenormalization,pairwisevariations
Vn/Vn+1werecalculatedbasedonthenormalizationfactor(NFnand
NFn+1)valuesaccordingtothegeNormalgorithm.IftheVn/Vn+1of ngeneswerebelowthecut-offvalue,theadditionalhousekeeping gene(n+1)wasnotnecessaryforreliablenormalization.The cut-offvalueofpairwisevariationsVn/Vn+1wassetat0.15.Intheresults
ofthesixgroupsanalyzed,atleastfourreferencegenescanbeused foraccuratenormalizationinAS(V2/V3=0.173,V3/V4=0.176),Y
(V2/V3=0.193,V3/V4=0.175)and F(V2/V3=0.193,V3/V4=0.183).
InW(V2/V3=0.22)andYF(V2/V3=0.203), thebestnumberwas
three.Twogenesweresufficientforexpressionnormalizationin WF(V2/V3<1.5)(Fig.4).
QuantificationofCtFAD2-10andCtKASIIexpressionwithstable referencegenes
TheexpressionofCtFAD2-10,oneoftheFAD2genefamilyin safflower,wasnormalizedusingKASIIinanearlystudy(Caoetal., 2013).Ketoacyl-acylcarrierproteinsynthaseII(KASII)alsoplays arolein thefattyacidbiosynthesisinplants.Theexpressionof
KASIIwasnotevaluatedforusingasaninternalcontrolfor nor-malization.Whencomparedwithorgansobtainedfromdifferent individuals,singlehousekeepinggeneRNAlevelswerenot appro-priatetobeusedfornormalizationofRNAlevels(Tricaricoetal., 2002).Tofurtherverifythesuitabilityofreferencegenesselected inthepresentstudy,CtFAD2-10andCtKASIIexpressionlevelswere detectedinsafflower(Fig.1).Therelativeexpressiondatawere cal-culatedusing2−Ctmethod.Theinternalcontrolgeneswerethe CtUBIandCt60SandthegeometricaverageofCtUBIandCt60S.
Itspattern of expressionwasassessed in allsamples(bract, ovary,stem,leaf,calyx,petal(I–IV)).Similarexpressionpatterns weregeneratedwheneitheroneortwoofthemoststablegenes wereusedfor normalization(Fig.1A and B).Whennormalized thegeometricaverageofCtUBIandCt60S,transcriptabundance ofCtKASIIgraduallyincreasedindifferentdevelopmentalstages offlower,peaking attheStageIV(Fig.1B).It suggeststhatthe
0.25
V2/3 V3/4 V4/5 V5/6 V6/7 V7/8 V8/9
0.20
0.15
0.10
0.05
0.00
AS Y W F YF WF
P
a
irwise v
a
riations
Fig.4. OptimalnumberofreferencegenesfornormalizationaccordingtogeNorm
results.Pairwisevariation(Vn/Vn+1)wasmeasuredbetweenthenormalization
fac-torsNFnandNFn+1.Theinclusionofanadditionalreferencegenewasnotrequired
belowthecut-offvalueof0.15(thechartreuseline).AS,allsamples;Y,ZHH0082
(Yellow);W,XinHonghuaNO.7(White).F,flower;YF,yellowflower;WF,white
Table3
RankingofthecandidatereferencegenesaccordingtotheirvaluescalculatedbygeNorm,NormFinder,CtapproachandBestKeeperinF,YFandWF.
Rank All flowers (F) Yellow-Flower (YF) White-Flower (WF)
geNorm(M) NormFinder (SV)
Ct
approach
BestKeeper geNorm(M) NormFinder (SV)
Ct
approach
BestKeeper geNorm(M) NormFinder (SV)
Ct
approach
BestKeeper
1 Ct60S CtACT Ct60S CtPP2A CtE1F4A CtE1F4A CtE1F4A CtPP2A Ct60S CtEF1 Ct60S CtPP2A
0.705 0.073 0.625 0.727 0.061 0.635 0.604 0.039 0.550
2 CtE1F4A CtEF1 CtACT CtUBI Ct60S CtACT Ct60S CtUBI CtE1F4A Ct60S CtACT CtUBI
0.742 0.086 0.650 0.755 0.071 0.665 0.657 0.050 0.583
3 CtUBI CtUBI CtE1F4A Ct60S CtTUB CtGAPDH CtTUB Ct60S CtACT CtPP2A CtTUB Ct60S
0.749 0.097 0.661 0.764 0.079 0.665 0.671 0.060 0.591
4 CtACT CtE1F4A CtUBI CtTUB CtUBI Ct60S CtACT CtE1F4A CtTUB CtUBI CtE1F4A CtTUB
0.777 0.097 0.665 0.768 0.098 0.687 0.678 0.060 0.593
5 CtTUA Ct60S CtGAPDH CtE1F4A CtGAPDH CtTUB CtUBI CtTUB CtTUA CtACT CtUBI CtE1F4A
0.806 0.100 0.707 0.819 0.109 0.692 0.683 0.091 0.594
6 CtGAPDH CtGAPDH CtTUB CtACT CtACT CtUBI CtGAPDH CtACT CtUBI CtTUA CtTUA CtGAPDH
0.808 0.112 0.709 0.829 0.110 0.697 0.687 0.098 0.637
7 CtTUB CtTUB CtTUA CtGAPDH CtEF1 CtEF1 CtEF1 CtEF1 CtPP2A CtE1F4A CtPP2A CtACT
0.809 0.123 0.754 0.836 0.130 0.767 0.745 0.126 0.666
8 CtEF1 CtPP2A CtPP2A CtTUA CtTUA CtTUA CtPP2A CtGAPDH CtGAPDH CtGAPDH CtGAPDH CtTUA
0.875 0.127 0.755 0.888 0.146 0.817 0.755 0.158 0.677
9 CtPP2A CtTUA CtEF1 CtEF1 CtPP2A CtPP2A CtTUA CtTUA CtEF1 CtTUB CtEF1 CtEF1
0.885 0.128 0.760 0.981 0.183 0.826 0.900 0.164 0.737
RankAggrega CtUBI/CtE1F4A/Ct60S CtE1F4A/Ct60S/CtTUB Ct60S/CtUBI/CtACT
aTopthreerankedgeneswereshown.
CtKASIIwasnotastablegeneforgeneexpressionnormalization. TheexpressionlevelsofCtFAD2-10werehighinpetals,especiallyin thefloweringstageIV(Fig.1A).Whenonlyonereferencegenewas employed,expressionprofilesofCtKASIIweresimilar(Fig.1B),but differenceswereevidentinestimatedtranscriptabundance,which washigherwhennormalizedagainstCtUBIthanagainstCt60S.
Discussion
Extensivestudiesarebeingcarriedoutonthesafflower,because itisbothanoilcropandamedicinalplant.Furthermore,research onthebiosynthesisoftheseflavonoidsandfattyacidmetabolism werealsoreportedrecently(Caoetal.,2013;Lietal.,2012).Because itisasensitive,specific,reproducible,andconventionalmethod, qPCR has become an essential tool for gene expression analy-sis,especially thegene expressionofthesecondary metabolites pathway(Al-Ghazietal.,2009).
Althoughsomereferencegeneshavebeenusedforthe normal-izationofgeneexpressioninsafflower,butthesereferencegenes werenotevaluated forqPCR datainsafflower tissues(Lietal., 2012;Caoetal.,2013).Recently,thesafflowerreferencegeneswere reportedinourprocessofrevising,buttheirtranscriptomedata wereonlyfromtheseedandalgorithmsforthereferencegenes evaluationwereincomplete(Lietal.,2015).Inthisstudy,two repre-sentativechemotypeofsafflowerlines,with-HSYA(yellowflower) andwithout-HSYA(whiteflower),waschosentoidentifythe saf-flowerreferencegenes.Theninehousekeepinggeneswereselected asthecandidategenesfromtranscriptomedataofdifferent devel-opmentalfloweringstageofsafflowerconstructedbyourlab.The profilingofthesegeneswascarriedoutusingfouralgorithmsand wasrankedbyRankAggreg.All54sampleswereseparatedintosix groupsinordertoevaluatethevariationofthesecandidategenesin differentlines,organs,andflowers.Althoughthereweredifferent listsofthemoststablyexpressedgenesfromtheresultsofgeNorm, NormFinder,CtapproachandBestKeeper,theCtUBIandCt60S
wereinthetopthreerankedgenesaccordingtotheRankAggreg analysisinthefivegroups(excepttheYF).Ourfindingswerein accordancewiththeresultthatUBQ(ubiquitin)showedhighly sta-bleexpressioninArabidopsis(Czechowskietal.,2005).Similarly,
UBQwastherecommendedhousekeepinggenefornormalization inpoplar(Brunneretal.,2004)andrice(Jainetal.,2006).However,
UBQwasnotsuggestedusingasinternalcontrolstonormalizegene expressioninsoybean(Jianetal.,2008).Asfor60S,itwasfoundto
bethebestreferencegeneindifferenttissuesandundervarious stressconditionsinsoybean(Leetal.,2012).Therefore,weused
CtUBIandCt60Sasreferencegenesandevaluatedtheexpression ofCtFAD2-10andCtKASIIinsafflower.CtKASIIhadbeenusedasthe referencegenetoprofiletheCtFAD2geneexpression.But accord-ingtoourstudy,theCtKASIIwasnotsuitableforgeneexpression normalizationbecauseitsexpressionswerehighlyvariable.
Ourresultsindicatedthatthestabilityofreferencegene expres-sionmustbevalidatedforeachlineandthedevelopmentstages offloweringinsafflower.Basedontheseresults,westrongly sug-gestthatCtUBIandCt60Sshouldbeusedasreferencegenesfor geneexpressioninsafflower.Theidentificationofthesetwostable referencegeneswillenableaccurateandreliablegeneexpression studiesrelated tofunctionalgenomicsandmetabolomics about safflower.
Ethicaldisclosures
Allauthorsdeclaretoourmanuscriptdidnotinvolveanyethical issues.
Authors’contribution
XYRandGDDwereresponsibleforthecollectionofplant sam-ple.LFandTYHdevelopedtheanalyticalmethodology.LFandGY wereresponsiblefordataanalysis.LFandGMLdesignedthestudy andwrotethemanuscript.Alltheauthorsreadthefinalmanuscript andapprovedthesubmission.
Conflictsofinterest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgments
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound,in theonlineversion,atdoi:10.1016/j.bjp.2016.05.006.
References
Al-Ghazi,Y.,Bourot,S.,Arioli,T.,Dennis,E.S.,Llewellyn,D.J.,2009.Transcript profil-ingduringfiberdevelopmentidentifiespathwaysinsecondarymetabolismand cellwallstructurethatmaycontributetocottonfiberquality.PlantCellPhysiol. 50,1364–1381.
Andersen,C.L.,Jensen,J.L.,Ørntoft,T.F.,2004.Normalizationofreal-time
quan-titativereversetranscription-PCR data:amodel-basedvarianceestimation approachtoidentifygenessuitedfornormalization,appliedtobladderandcolon cancerdatasets.CancerRes.64,5245–5250.
Andersen,O.M.,Markham,K.R.,2010.Flavonoids:Chemistry,Biochemistryand
Applications.CRCPress.
Asgarpanah,J.,Kazemivash,N.,2013.Phytochemistry,pharmacologyandmedicinal
propertiesofCarthamustinctoriusL.Chin.J.Integr.Med.19,153–159.
Bas, A., Forsberg, G., Hammarström, S., Hammarström, M.L., 2004. Utility
of the housekeeping genes 18S rRNA, -Actin and glyceraldehyde-3-phosphate-dehydrogenasefornormalizationinreal-timequantitativereverse transcriptase-polymerasechainreactionanalysisofgeneexpressioninhuman Tlymphocytes.Scand.J.Immunol.59,566–573.
Brunner,A.M.,Yakovlev,I.A.,Strauss,S.H.,2004.Validatinginternalcontrolsfor
quantitativeplantgeneexpressionstudies.BMCPlantBiol.4,14.
Bustin,S.A.,Benes,V.,Garson,J.A.,Hellemans,J.,Huggett,J.,Kubista,M.,Mueller,R., Nolan,T.,Pfaffl,M.W.,Shipley,G.L.,Vandesompele,J.,Wittwer,C.T.,2009.The MIQEguidelines:minimuminformationforpublicationofquantitative real-timePCRexperiments.Clin.Chem.55,611–622.
Cao,S.,Zhou,X.R.,Wood,C.C.,Green,A.G.,Singh,S.P.,Liu,L.,Liu,Q.,2013.Alarge andfunctionallydiversefamilyofFad2genesinsafflower(Carthamustinctorius
L.).BMCPlantBiol.13,5.
Chapman,M.,Burke,J.,2007.DNAsequencediversityandtheoriginofcultivated
safflower(CarthamustinctoriusL.;Asteraceae).BMCPlantBiol.7,60.
Chapman,M.A.,Hvala,J.,Strever,J.,Matvienko,M.,Kozik,A.,Michelmore,R.W.,Tang,
S.,Knapp,S.J.,Burke,J.M.,2009.Development,polymorphism,andcross-taxon
utilityofEST–SSRmarkersfromsafflower(CarthamustinctoriusL.).Theor.Appl. Genet.120,85–91.
Czechowski,T.,Stitt,M.,Altmann,T.,Udvardi,M.K.,Scheible,W.-R.,2005.
Genome-wideidentification and testing of superior reference genes for transcript normalizationinArabidopsis.PlantPhysiol.139,5–17.
Feng,N.,Li,Y.,Tang,J.,Wang,Y.,Guo,M.,2010.cDNA-AFLPanalysison
trans-criptsassociatedwithhydroxysaffloryellowA(HSYA)biosyntheticpathwayin
Carthamustinctorius.Biochem.Syst.Ecol.38,971–980.
Feng,Z.M.,He,J.,Jiang,J.S.,Chen,Z.,Yang,Y.N.,Zhang,P.C.,2013.NMRsolution
structurestudyoftherepresentativecomponenthydroxysaffloryellowAand otherquinochalconeC-glycosidesfromCarthamustinctorius.J.Nat.Prod.76, 270–274.
Gachon,C.,Mingam,A.,Charrier,B.,2004.Real-timePCR:whatrelevancetoplant
studies?J.Exp.Bot.55,1445–1454.
Gecgel,U.,Demirci,M.,Esendal,E.,Tasan,M.,2007.Fattyacidcompositionoftheoil fromdevelopingseedsofdifferentvarietiesofsafflower(Carthamustinctorius
L.).J.Am.OilChem.Soc.84,47–54.
Golkar,P.,Arzani,A.,Rezaei,A.M.,2011.Geneticvariationinsafflower(Carthamus
tinctoriousL.)forseedquality-relatedtraitsandinter-simplesequencerepeat (ISSR)markers.Int.J.Mol.Sci.12,2664–2677.
Guénin,S.,Mauriat,M.,Pelloux,J.,VanWuytswinkel,O.,Bellini,C.,Gutierrez,L.,2009.
NormalizationofqRT-PCRdata:thenecessityofadoptingasystematic, experi-mentalconditions-specific,validationofreferences.J.Exp.Bot.60,487–493.
Guan,L.,Wu,W.,Hu,B.,Li,D.,Chen,J.,Hou,K.,Wang,L.,2014.Devolopmental
andgrowthtemperatureregulationofomega-3fattyaciddesaturasegenesin safflower(CarthamustinctoriusL.).Genet.Mol.Res.13,6623.
Gutierrez,L.,Mauriat,M.,Guénin,S.,Pelloux,J.,Lefebvre,J.-F.,Louvet,R.,Rusterucci,
C.,Moritz,T.,Guerineau,F.,Bellini,C.,VanWuytswinkel,O.,2008.Thelack
ofasystematicvalidationofreferencegenes:aseriouspitfallundervaluedin reversetranscription-polymerasechainreaction(RT-PCR)analysisinplants. PlantBiotechnol.J.6,609–618.
Infante,C.,Matsuoka,M.,Asensio,E.,Canavate,J.,Reith,M.,Manchado,M.,2008.
Selectionofhousekeepinggenesforgeneexpressionstudiesinlarvaefrom flatfishusingreal-timePCR.BMCMol.Biol.9,28.
Jain,M.,Nijhawan,A.,Tyagi,A.K.,Khurana,J.P.,2006.Validationofhousekeeping
genesasinternalcontrolforstudyinggeneexpressioninricebyquantitative real-timePCR.Biochem.Biophys.Res.Commun.345,646–651.
Jarosova,J.,Kundu,J.,2010.Validationofreferencegenesasinternalcontrolfor
studyingviralinfectionsincerealsbyquantitativereal-timeRT-PCR.BMCPlant Biol.10,146.
Jian,B.,Liu,B.,Bi,Y.,Hou,W.,Wu,C.,Han,T.,2008.Validationofinternalcontrolfor geneexpressionstudyinsoybeanbyquantitativereal-timePCR.BMCMol.Biol. 9,59.
Jun,M.S.,Ha,Y.M.,Kim,H.S.,Jang,H.J.,Kim,Y.M.,Lee,Y.S.,Kim,H.J.,Seo,H.G.,Lee,
J.H.,Lee,S.H.,Chang,K.C.,2011.Anti-inflammatoryactionofmethanolextract
ofCarthamustinctoriusinvolvesinhemeoxygenase-1induction.J. Ethnophar-macol.133,524–530.
Le,D.T.,Aldrich,D.L.,Valliyodan,B.,Watanabe,Y.,VanHa,C.,Nishiyama,R.,
Gut-tikonda,S.K.,Quach,T.N.,Gutierrez-Gonzalez,J.J.,Tran,L.-S.P.,2012.Evaluation ofcandidatereferencegenesfornormalizationofquantitativeRT-PCRin soy-beantissuesundervariousabioticstressconditions.PLoSONE7,e46487.
Li,D.,Hu,B.,Wang,Q.,Liu,H.,Pan,F.,Wu,W.,2015.Identificationandevaluation ofreferencegenesforaccuratetranscriptionnormalizationinsafflowerunder differentexperimentalconditions.PLOSONE10,e0140218.
Li,H.,Dong,Y.,Yang,J.,Liu,X.,Wang,Y.,Yao,N.,Guan,L.,Wang,N.,Wu,J.,Li,X.,2012. Denovotranscriptomeofsafflowerandtheidentificationofputativegenesfor oleosinandthebiosynthesisofflavonoids.PLoSONE7,e30987.
Li,Y.,Wang,Z.,Chang,H.,Wang,Y.,Guo,M.,2010.ExpressionofCT-wpr,screened
bycDNA-AFLPapproach,associatedwithhydroxysaffloryellowAinCarthamus tinctoriusL.Biochem.Syst.Ecol.38,1148–1155.
Liu,D.,Shi,L.,Han,C.,Yu,J.,Li,D.,Zhang,Y.,2012.Validationofreferencegenesfor geneexpressionstudiesinvirus-infectedNicotianabenthamianausing quanti-tativereal-timePCR.PLoSONE7,e46451.
Liu,Q.,Cao,S.,Zhou,X.-R.,Wood,C.,Green,A.,Singh,S.,2013.Nonsense-mediated
mRNAdegradationofCtFAD2-1anddevelopmentofaperfectmolecularmarker forololmutationinhigholeicsafflower(CarthamustinctoriusL.).Theor.Appl. Genet.126,2219–2231.
Mallona,I.,Lischewski,S.,Weiss,J.,Hause,B.,Egea-Cortines,M.,2010.Validationof referencegenesforquantitativereal-timePCRduringleafandflower develop-mentinPetuniahybrida.BMCPlantBiol.10,4.
Peng,S.,Feng,N.,Guo,M.,Chen,Y.,Guo,Q.,2008.GeneticvariationofCarthamus
tinctoriusL.andrelatedspeciesrevealedbySRAPanalysis.Biochem.Syst.Ecol. 36,531–538.
Pfaffl,M.W.,Tichopad,A.,Prgomet,C.,Neuvians,T.P.,2004.Determinationofstable
housekeepinggenes,differentiallyregulatedtargetgenesandsampleintegrity: BestKeeper–Excel-basedtoolusingpair-wisecorrelations.Biotechnol.Lett.26, 509–515.
Pihur,V.,Datta,S.,Datta,S.,2009.RankAggreg,anRpackageforweightedrank
aggregation.BMCBioinform.10,62.
Ruijter,J.M.,Ramakers,C.,Hoogaars,W.M.H.,Karlen,Y.,Bakker,O.,vandenHoff,
M.J.B.,Moorman,A.F.M.,2009.Amplificationefficiency:linkingbaselineandbias intheanalysisofquantitativePCRdata.NucleicAcidsRes.37,e45.
Salem,N.,Msaada,K.,Hamdaoui,G.,Limam,F.,Marzouk,B.,2011.Variationin
phenoliccompositionandantioxidantactivityduringflowerdevelopmentof safflower(CarthamustinctoriusL.).J.Agric.FoodChem.59,4455–4463.
Schmidt,G.W.,Delaney,S.K.,2010.Stableinternalreferencegenesfornormalization
ofreal-timeRT-PCRintobacco(Nicotianatabacum)duringdevelopmentand abioticstress.Mol.Genet.Genomics283,233–241.
Schmittgen,T.D.,Livak,K.J.,2008.Analyzingreal-timePCRdatabythecomparative
CTmethod.Nat.Protoc.3,1101–1108.
Silveira,E.,Alves-Ferreira,M.,Guimaraes,L.,daSilva,F.,Carneiro,V.,2009. Selec-tionofreferencegenesforquantitativereal-timePCRexpressionstudiesinthe apomicticandsexualgrassBrachiariabrizantha.BMCPlantBiol.9,84.
Silver,N.,Best,S.,Jiang,J.,Thein,S.,2006.Selectionofhousekeepinggenesforgene expressionstudiesinhumanreticulocytesusingreal-timePCR.BMCMol.Biol. 7,33.
Sun,L.,Yang,L.,Fu,Y.,Han,J.,Xu,Y.,Liang,H.,Cheng,Y.,2013.CapacityofHSYA toinhibitnitrotyrosineformationinducedbyfocalischemicbraininjury.Nitric Oxide35,144–151.
Tanaka,Y.,Brugliera,F.,Kalc,G.,Senior,M.,Dyson,B.,Nakamura,N.,Katsumoto,Y.,
Chandler,S.,2010.Flowercolormodificationbyengineeringoftheflavonoid
biosyntheticpathway:practicalperspectives.Biosci.Biotechnol.Biochem.74, 1760–1769.
Thellin,O.,Zorzi,W.,Lakaye,B.,DeBorman,B.,Coumans,B.,Hennen,G.,Grisar,T.,
Igout,A.,Heinen,E.,1999.Housekeepinggenesasinternalstandards:useand
limits.J.Biotechnol.75,291–295.
Tian,Y.,Yang,Z.-F.,Li,Y.,Qiao,Y.,Yang,J.,Jia,Y.-Y.,Wen,A.-D.,2010.Pharmacokinetic comparisonsofhydroxysaffloweryellowAinnormalandbloodstasissyndrome rats.J.Ethnopharmacol.129,1–4.
Tricarico,C.,Pinzani,P.,Bianchi,S.,Paglierani,M.,Distante,V.,Pazzagli,M.,Bustin,
S.A.,Orlando,C.,2002.Quantitativereal-timereversetranscriptionpolymerase
chainreaction:normalizationtorRNAorsinglehousekeepinggenesis inappro-priateforhumantissuebiopsies.Anal.Biochem.309,293–300.
Vandesompele,J.,DePreter,K.,Pattyn,F.,Poppe,B.,VanRoy,N.,DePaepe,A.,
Speleman,F.,2002.Accuratenormalizationofreal-timequantitativeRT-PCR
databygeometricaveragingofmultipleinternalcontrolgenes.GenomeBiol.3, 1–12.
Wan,H.,Yuan,W.,Ruan,M.,Ye,Q.,Wang,R.,Li,Z.,Zhou,G.,Yao,Z.,Zhao,J.,Liu,S.,
Yang,Y.,2011.Identificationofreferencegenesforreversetranscription
quan-titativereal-timePCRnormalizationinpepper(CapsicumannuumL.).Biochem. Biophys.Res.Commun.416,24–30.
Wang,C.,Huang,Q.,Zhu,X.,Duan,Y.,Yuan,S.,Bai,X.,2013.Hydroxysaffloryellow
Asuppressesoleicacid-inducedacutelunginjuryviaproteinkinaseA.Toxicol. Appl.Pharmacol.272,895–904.
Yang,J.,Wang,Y.,Guo,M.-L.,2011.Identificationandmappingofanovel
hydrox-ysaffloryellowA(HSYA)biosyntheticgeneinCarthamustinctorius.Biochem. Genet.49,410–415.
Yeilaghi,H.,Arzani,A.,Ghaderian,M.,Fotovat,R.,Feizi,M.,Pourdad,S.S.,2012.Effect ofsalinityonseedoilcontentandfattyacidcompositionofsafflower(Carthamus tinctoriusL.)genotypes.FoodChem.130,618–625.