___________________________
Corresponding author: Sanjay Kumar, Genetics Laboratory, Department of Zoology, Banaras
Hindu University, Varanasi 225001, India (Phone: +91 7860448788; E-mail:
sanjay_mbt@rediffmail.com)
UDC 575:633 DOI: 10.2298/GENSR1403681K
Original scientific paper
STUDIES ON EFFICIENCY OF RAPD PRIMERS IN DEVELOPING MOLECULAR PROFILES FOR GENETIC PURITY STUDIES IN SOYBEAN (Glycine max L.)
CULTIVARS
Sanjay KUMAR
Genetics Laboratory, Department of Zoology, Banaras Hindu University, Varanasi, India
Kumar S. (2014): Studies on efficiency of RAPD primers in developing molecular profiles for genetic purity studies in soybean (Glycine max L.) cultivars. - Genetika, Vol 46, No. 3, 681- 692.
Major advances have recently been made in our understanding of soybean genetics and of the application of new technologies to soybean improvement. Estimates of genetic relationships on the basisof the enzymes and molecular markers have been shown to be consistentwith expectations based on origin and pedigree information. To identify efficient markers, that are to be used for genetic purity studies, polymorphism is the basic criterion. RAPD has been found to be an effective and efficient tool to evaluate and reveal genetic polymorphism in several crop species. In present study a total of 80 RAPD primers were screened, out of which 37 gave amplification and only 30 primers showed unambiguous DNA profile. Out of these 30 primers, 22 gave polymorphic banding patterns. It is evident from the result that, 30/80 primers tried (38%) provided unambiguous amplification and out of 30 primers, 22 primers (73%) were found to be polymorphic. Scorable 30 RAPD primers led to amplification of 120 fragments out of which 81 (67.5%) bands were found to be polymorphic. On an average, we got 4 bands per primer and 15 primers (50%) have been found to produce more number of bands than the average value which is encouraging. 8 primers were found to give 100% polymorphisms. Our results are indicative of the efficiency of RAPD primers towards development of molecular profiles.
Key words:Glycine max, Polymorphism, PCR, RAPD, UPGMA
INTRODUCTION
soil-climatic conditions. Being one of the main oil crops of the world, in India it is grown in all its major states viz. Madhya Pradesh, Uttar Pradesh, Maharashtra, Rajasthan and Gujarat. It is also grown on a small scale in Himachal Pradesh, Punjab and Delhi. Major advances have recently been made in our understanding of soybean genetics and of the application of new technologies to soybean improvement. Thus it is now possible, using molecular methods, to alter the protein and oil composition of soybean, as well as to produce other foreign proteins in the plant. Traditionally, the study of genetic diversity has fallen within population genetics which has focussed on measuring its extent in natural populations, in comparing levels of genetic diversity within and among populations and in making references on the nature and intensity of evolutionary processes from the observed patterns of genetic diversity. Hence, there is a long tradition as well as a wealth of conceptual tools in population genetics for analyzing, measuring and partitioning genetic diversity (KUMAR and SINGH 2014a; b).
Several thousand soybean accessions from the USDA Soybean GermplasmCollection (Urbana, IL) have been evaluated for agronomic andseed composition traits as well as disease resistance (NELSON et al., 1987; NELSON et al., 1989; JUVIK et al., 1989; BERNARD et al., 1989) although a large number of G. max lines are availableto soybean breeding programs, discovering and transferring novel,favourable genes from G. soja into G. max could be a successfulapproach for enhancing the genetic variability and improvingcultivated soybean (CARPENTER and FEHR, 1986). Identifying useful diversity is a challenge. Molecular characterization of soybean germplasm has been performed with isozymes, proteins, andDNA markers. Estimates of genetic relationships on the basisof the enzymes and molecular markers have been shown to be consistent with expectations based on origin and pedigree information (GRIFFIN and PALMER, 1995; MAUGHAN et al., 1995, 1996; DOLDI et al., 1997; THOMPSON et al., 1998). KEIM et al. (1989) surveyed 58 G. max and G. soja accessionswith 17 restriction fragment length polymorphism (RFLP) markersand found that the molecular diversity was the least among cultivatedsoybeans and greatest between species. Using simple sequencerepeat (SSR) and amplified sequence length polymorphism (ASLP)markers MAUGHAN et al. (1995) reported that five microsatellitemarkers detected a total of 79 alleles in a sample of 94 accessionsof wild and cultivated soybean. Allelic diversity for the SSRloci was greater in wild soybean than in cultivated soybean.Overall, 43 more SSR alleles were detected in wild than in cultivatedsoybean. MAUGHAN et al. (1996) evaluated 23 accessions of wildand cultivated soybean using amplified fragment length polymorphism(AFLP). Among the 759 AFLP fragments scored, 17% were polymorphic in G. max and 31% were polymorphic in G. soja. Their resultsalso indicated that AFLP phenotypic variation was greater in wild soybean than in cultivated soybean. GRIFFIN and PALMER (1995)screened more than 1200 G. max and G. soja plant introductionswith eight enzymes. The numbers of alleles per locus and averagegene diversity were greater in the G. soja samples than in theG. max samples.
MATERIALS AND METHODS Plant material
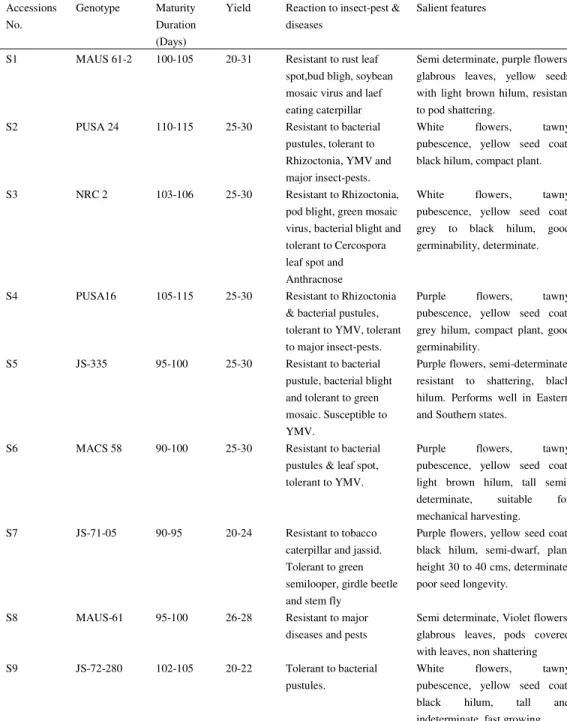
Table 1. Name, parentage and morphological descriptions of the soybean varieties used in the present study
Accessions No.
Genotype Maturity Duration (Days)
Yield Reaction to insect-pest & diseases
Salient features
S1 MAUS 61-2 100-105 20-31 Resistant to rust leaf spot,bud bligh, soybean mosaic virus and laef eating caterpillar
Semi determinate, purple flowers, glabrous leaves, yellow seeds with light brown hilum, resistant to pod shattering.
S2 PUSA 24 110-115 25-30 Resistant to bacterial
pustules, tolerant to Rhizoctonia, YMV and major insect-pests.
White flowers, tawny pubescence, yellow seed coat, black hilum, compact plant.
S3 NRC 2 103-106 25-30 Resistant to Rhizoctonia,
pod blight, green mosaic virus, bacterial blight and tolerant to Cercospora leaf spot and Anthracnose
White flowers, tawny pubescence, yellow seed coat, grey to black hilum, good germinability, determinate.
S4 PUSA16 105-115 25-30 Resistant to Rhizoctonia
& bacterial pustules, tolerant to YMV, tolerant to major insect-pests.
Purple flowers, tawny pubescence, yellow seed coat, grey hilum, compact plant, good germinability.
S5 JS-335 95-100 25-30 Resistant to bacterial
pustule, bacterial blight and tolerant to green mosaic. Susceptible to YMV.
Purple flowers, semi-determinate, resistant to shattering, black hilum. Performs well in Eastern and Southern states.
S6 MACS 58 90-100 25-30 Resistant to bacterial
pustules & leaf spot, tolerant to YMV.
Purple flowers, tawny pubescence, yellow seed coat, light brown hilum, tall semi-determinate, suitable for mechanical harvesting.
S7 JS-71-05 90-95 20-24 Resistant to tobacco
caterpillar and jassid. Tolerant to green semilooper, girdle beetle and stem fly
Purple flowers, yellow seed coat, black hilum, semi-dwarf, plant height 30 to 40 cms, determinate, poor seed longevity.
S8 MAUS-61 95-100 26-28 Resistant to major
diseases and pests
Semi determinate, Violet flowers, glabrous leaves, pods covered with leaves, non shattering S9 JS-72-280 102-105 20-22 Tolerant to bacterial
pustules.
S10 KALITURE 120-130 18-20 Susceptible to soybean mosaic, tolerant to bacterial pustules
Purple flowers, tawny pubescen, black seed coat, black hilum, small seeded semi indeterminate S11 PUNJAB 1 95-100 25-30 Susceptible to bacterial
pustules, less susceptible to blue beetle.
Purple flowers, yellow pods with brown pubescence, yellow seed coat, small seeds, light brown hilum, high pod-shattering, early, good germinability, suitable for food uses.
S12 INDIRA
SOYA 9
106 22-23 Resistant to rust. Moderately resistant to stem tunneling and girdle beetle and leaf folder. Performs well under low to moderate plant densities
Light grey pubescence throughout the plant parts, broad light medium size green leaves, yellow seeds of medium size with black hilum and intermediate lustre.
S13 JS-80-21 105-110 25-30 Tolerant to bacterial pustules, viral diseases and foliar insect-pests.
Purple flowers, tawny pubescence, yellow seed coat, brown/black hilum, determinate, high seed germinability. Perform well in eastern states.
S14 JS-2 90-95 18-20 Resistant to bacterial
pustule, tolerant to Macrophomina
Purple flowers, tawny pubescence, pods with dense brown pubescene, yellow seed coat, light brown hilum, determinate, highly shattering
S15 MAUS 71 93-100 18-30 Not known Semi determinate, purple flowers,
glabrous leaves, yellow seed with black hilum, resistant to pod shattering.
S16 SHYAMA 105-110 20-25 Resistant to bacterial
pustules and seed and seed ling rot, tolerant to Anthracnose
Purple flowers, brown pubescen, black seed coat, buff coloured hilum, medium tall plants, suited to low input conditions. S17 MACS 124 95-105 25-30 Resistant to bud blight,
soybean mosaic and bacterial pustules.
Purple flowers,
tawny pubescence, yellow seed coat, dark brown hilum, semi-determinate resistant to lodging. S18 MACS 450 90-95 25-40 Resistant to leaf spot,
bud blight, yellow mosaic, soybean mosaic, bacterial pustule. Highly resistant to stemfly and defoliators.
S19 BRAGG 112-115 15-20 Resistant to bacterial pustules, susceptible to YMV.
White flowers, grey pubescence, yellow seed coat, black hilum, brown pods, determinate.
S20 PK 416 115-120 30-35 Resistant to YMV &
bacterial pustules, tolerant to Rhizoctonia.
White flowers, tawny pubescence, yellow seed coat,
brown hilum,
semi-determinate. Performs well in Punjab, Haryana and also in central zone.
S21 SL 295 120-130 20-25 Resistant to yellow
mosaic virus
White flower, tawny pubescence, determinate, yellow seed coat, black hilum.
S22 NRC 37 96-102 35-40 Moderately resistant to
collar rot, bacterial pustule, pod blight and bud blight like syndrome. Moderately resistant to stem fly and leaf miner. Non lodging under optimum plant population, non shattering behaviour upto 10 days after harvest maturity
Erect, determinate plants with out anthocyanin colouration in the hypocotyl, white flowers, tawny pubescence, small to medium, spherical yellow seeds with light to dark brown hilum.
S23 PUSA 20 105-120 25-30 Resistant to Rhizoctonia & bacterial pustules, tolerant to YMV and major insect-pests.
White flowers, tawny pubescence, yellow seed coat, black hilum, compact plant. Performs well in central zone.
S24 NRC 7 90-99 25-35 Resistant to bacterial
blight, green mosaic virus, bacterial pustules, phyllody, soybean mosaic, Myrothe- cium and Cercospora leaf spots, tolerant to stem fly, girdle beetle, green and grey semilooper, leaf miner and defoliators.
Determinate, grey pubescence, purple flowers, yellow seed coat, brown hilum, high oil content, resistant to pod-shattering.
Isolation of genomic DNA
10,000 rpm for 10 min. The upper aqueous phase was extracted 2-3 times with fresh chloroform: iso-amyl alcohol (24:1). The final aqueous phase was transferred to other centrifuge tubes. To these, 0.6 volume of ice cold iso-propanol was added and mixed gently by inverting the tube. DNA complex was spooled out with a bent pasture pipette and 20 ml of washing solution was added to it. The pellet was gently agitated for few minutes and collected by centrifugation at 40C, dried and an appropriate volume of TE buffer was used to dissolve the pellet. Further the RNaseA @ 10µg/ml was added into the DNA which was then incubated at 370C for 30 minutes. Equal volume of phenol: chloroform: iso-amyl alcohol (25:24:1) was added into it and centrifuged at 10000 rpm for10 minutes. Aqueous phase was taken and equal volumes of chloroform: iso-amyl alcohol (24:1) was added and centrifuged at 10000 rpm for 10 min. To the aqueous phase, 1/20th volume of sodium acetate (3M, pH5.2) and 2.5 volume of ethanol was added. Next it was incubated at -200C for 1h (or-700C for 30 min). Then the solution was centrifuged at 10000 rpm for 10 min. Thereafter, pellets were washed with 70% ethanol (10000 rpm, 5 min), air dried and dissolved in distilled water. Yield of DNA was estimated using DNA markers with known DNA – uncut, through electrophoresis.
PCR amplification and gel electrophoresis
The random amplification was performed following a modified method of WILLIAMS et al. (1990). The reaction was carried out in a thermal cycle (Mastercycler, Eppendorf). A total of 80
RAPD primers were screened, out of which 37 produced amplifications and only 30 primers (viz. OPA 01, 03, 04, 08, 13 ; OPJ 01, 05, 09, 11, 12, 13, 16, 17, 18, 19; OPP 03, 04, 05, 06, 07, 08, 09 and OPT 01, 05, 07, 12, 13, 15, 16, 20, Operon Technologies, Canada, USA) showed unambiguous DNA profile (Table 2). Polymerase chain reaction mixture of 25 µl contained 25 ng of genomic DNA template, 0.6 U of Taq DNA polymerase (Bangalore Genei, Bangalore, India), 0.3 M decamer primer (Operon Technologies, Alameda, CA, USA), 2.5 µl 10 X PCR assay buffer (50 mM KCl, 10 mM Tris–HCl, 1.5 mM MgCl2) and 0.25 µ1 pooled dNTPs (100 mM each of dATP, dCTP, dGTP and dTTP from Promega, USA). PCR cycle conditions were as follows: initial denaturing step at 94°C for 3 min followed by 44 cycles of 94°C for 1 min, 37°C for 1 min and 72°C for 2 min. In the last cycle, primer extension at 72°C for 7 min was provided. PCR products separation was done through 1.5% agarose gel electrophoresis alongside O’Gene RulerTM 100 bp DNA Ladder Plus (Fermentas Life Sciences) as molecular mass marker. The amplified products were documented under UV light source.
Data analysis
Table 2. Name, sequence and details of the polymorphic RAPD primers used in the present study
RESULTS
Out of total 80 primers (Kits OPA, OPP, OPT and OPJ) tried in this study 37 primers showed clear and unambiguous amplification (Table 3). Scorable 30 RAPD primers led to amplification of 120 fragments ranging from about 3,300 bp (OPT 1) to 300 bp (OPT 12), out of which 81 (67.5%) bands were found to be polymorphic. The level of polymorphism ranged from 33.4% (OPT 16, where 1/3 bands was found to be polymorphic) to 100% (OPA1, 4, OPP 5, 7, 8, 9, 15 and OPT 13). Maximum number of 7 amplified products were obtained by primer OPT 01 and 13. Eight primers were found to be monomorphic and the primer OPA 03 amplified a
Primer Sequence (5'-3') Molecular weight
No. of bands amplified
No. of polymorphic
band
% polymorphism
OPA-01 CAGGCCCTTC 2964 6 6 100
OPA-03 AGTCAGCCAC 2997 1 0 0
OPA-04 AATCGGGCTG 3068 5 5 100
OPA-08 GTGACGTAGG 3108 2 1 50
OPA-13 CAGCACCCAC 2942 5 2 40
OPJ-01 CCCGGCATAA 2997 2 1 50
OPJ-05 CTCCATGGGG 3044 2 0 0
OPJ-09 TGAGCCTCAC 2988 1 0 0
OPJ-11 ACTCCTGCGA 2988 6 3 50
OPJ-12 GTCCCGTGGT 3035 5 4 80
OPJ-13 CCACACTACC 2917 3 1 33.3
OPJ-16 CTGCTTAGGG 3059 1 0 0
OPJ-17 ACGCCAGTTC 2988 3 0 0
OPJ-18 TGGTCGCAGA 3068 5 5 100
OPJ-19 GGACACCACT 2997 3 2 66.67
OPP-03 CTGATACGCC 2988 5 2 40
OPP-04 GTGTCTCAGG 3059 1 0 0
OPP-05 CCCCGGTAAC 2973 5 5 100
OPP-06 GTGGGCTGAC 3084 5 4 80
OPP-07 GTCCATGCCA 2988 4 4 100
OPP-08 ACATCGCCCA 2957 7 7 100
OPP-09 GTGGTCCGCA 3044 4 4 100
OPT-01 GGGCCACTCA 3013 7 6 85.7
OPT-05 GGGTTTGGCA 3099 3 0 0
OPT-07 GGCAGGCTGT 3084 5 2 40
OPT-12 GGGTGTGTAG 3139 5 4 80
OPT-13 AGGACTGCCA 3037 7 7 100
OPT-15 GGATGCCACT 3028 6 5 83.3
OPT-16 GGTGAACGCT 3068 3 1 33.3
minimum of 1 band. On an average we obtained 4 bands per primer and 22 primers used in the study produced polymorphic banding pattern. RAPD product (750 bp) produced by OPT 07 was found to be specific for MAUS-61 , while OPJ 18 produced a 2350 bp band only in Indira Soya 9. DNA amplification pattern as detected by some of the RAPD primers in the soybean cultivars has been provided in Fig. 1.
Figure 1.RAPD profile of soybean varieties obtained with primers OPA-04, OPJ-12 and OPT-13. Serial number of the varieties corresponds to Table 1. M = Standard DNA marker, 100 bp DNA Ladder Plus
OPA-04
M 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24
OPJ-12
M 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24
OPT-13
Table 3. Similarity matrix of 24 soybean cultivars
Thus, it is evident that, 30/80 primers tried (38%) provided unambiguous amplification and out of 30 primers, 22 primers (73%) were found to be polymorphic.
DISSCUSSION
RAPD has been found to be an effective and efficient tool to evaluate and reveal genetic polymorphism in several crop species like rice (VIRK et al., 1995; RAY-CHOUDHURY et al., 2001), wheat (CAO et al., 2000), maize (PEJIC et al., 1998), barley (YU et al., 2002) and soybean (BARANEK et al., 2002; BARAKAT, 2004). In our present study 67.5% fragments were found to be polymorphic as compared to 46% obtained by BARANEK et al. (2002) in 19 Czech National Collection of soybean genotypes. Further, THOMPSON et al. (1998) were able to identify only 34% polymorphism using RAPD. Very high degree of polymorphism detected in our present study indicated an accurate selection of the polymorphic RAPD primers. The 30 RAPD primers used in the present study has been selected after screening of 80 primers, 22 out of 30 were found to be polymorphic. Thus, quite a high percentage of primers (73%, as mentioned earlier). Two genotypes, were able to generate specific unique band. However, validation of the identified primers and reproducibility of those bands are required for proper identification of the genotypes. Moreover, the unique bands could be converted into SCAR (Sequence Characterized Amplified Region) markers for specificity. In this regard, few more RAPD primers could be tried for identification of unique bands in newer varieties. On an average, we got 4 bands per primer. Further, 8 primers were found to give 100% polymorphism among the genotypes studied. 15 primers (50%) have been found to develop more number of bands than the average value which is encouraging (Table 2) and indicates the efficiency of RAPD primers towards development of molecular profiles.
ACKNOWLEDGEMENTS
Author is very grateful to Directorate of Seed Research, Kushmaur, Mau. India, to provide facilities for this work. Special thanks are due to Dr. P. RAY-CHOUDHURY and Mr. S.P. TRIPATHI, for his guidance during this work.
Received December 16th
, 2013 Accepted September 05th
, 2014
REFERENCES
BARAKAT, H. (2004): Genetic fingerprinting and relationships of six soybeans (Glycine max L.) cultivars based on protein and DNA polymorphism. Int. J. Agri. and Biol., 6: 877-883.
BARANEK, M., M. KADLEC, J. RADDOVA, M. VACHUN and M. PIDRA (2002): Evaluation of genetic diversity in 19 Glycine max (L.) Merr. accessions included in the Czech national collection of soybean genotypes. Czech. J. Genet. Plant Breed, 38: 69–74.
BERNARD, R.L., G.A. JUVIK and R.L. NELSON (1989): USDA Soybean Germplasm Collection Inventory. Int. Agri. Pub., Urbana, IL.
CAO, W., G. SCOLES, P. HUCL and R.N. CHIBBAR (2000): Phylogenetic relationships of five morphological groups of hexaploid wheat (Triticum aestivum L.) based on RAPD analysis. Genome, 43: 724–727.
CARPENTER, J.A. and W.R. FEHR (1986): Genetic variability for desirable agronomic traits in populations containing Glycine soja germplasm. Crop Sci., 26: 681–686.
GRIFFIN, J.D. and R.G. PALMER (1995): Variability of thirteen isozyme loci in the USDA soybean germplasm collections. Crop Sci., 35: 897–904.
KUMAR, S. and A.K. SINGH (2014a): Complete absence of linkage disequilibrium between enzyme loci in natural populations of Drosophila ananassae. Genetika 46: 227–234.
KUMAR, S. and A.K. SINGH (2014b): Allozyme polymorphism in Drosophila. Proc. Zool. Soc. 67: (in press) DOI 10.1007/s12595-014-0126-3
JUVIK, G., R.L. BERNARD, R.Z. CHANG and J.F. CAVINS (1989): Evaluation of the USDA Wild Soybean Germplasm Collection: Maturity Group 000 to IV (PI 65.549 to PI 483.464). U.S. Department of Agriculture. Tech Bull 1761, U.S. Govt Print Office, Washington, DC.
KEIM, P., R.C. SHOEMAKER and R.G. PALMER (1989): Restriction fragment length polymorphism diversity in soybean. Theor. Appl. Genet., 77: 786–797.
MAUGHAN, P.J., M.M.A. SAGHAI and G.R. BUSS (1996): Molecular-marker analysis of seed-weight: Genomic locations, gene action, and evidence for orthologous evolution among three legume species. Theor. Appl. Genet., 93: 574–579.
MAUGHAN, P.J., M.M.A. SAGHAI and G.R. BUSS (1995): Microsatellite and amplified sequence length polymorphisms in cultivated and wild soybean. Genome, 38: 715-723.
MURRAY, M.G. and W.F. THOMSON (1980): Rapid isolation of high molecular weight plant DNA. Nucl. Acid Res., 8: 4321-4325.
NELSON, A.I., W.B. WIJERATNE, S.W. YEN, T.M. WEI and L.S. WEI (1987): Dry extrusion as an aid to mechanical expelling of oil from soybeans. J. Am. Oil. Chem. Soc., 64: 1341–1347.
NELSON, R.L., C.D. NICKELL, J.H. ORF, H. TACHIBANA, E.T. GRITTON, C.R. GRAU and B.W. KENNEDY (1989): Evaluating Soybean Germ Plasm for Brown Stem Rot Resistance. Plant Disease, 73: 110-114.
PEJIC, I., P. AJMONE-MARSON, M. MORGANTE, V. KOZUMPLICK, P. CASTIGLIONI, G. TARAMINO and M. MOTT (1998): Comparative analysis of genetic similarity among maize inbred lines detected by RFLPs, RAPDs, SSRs and AFLPs. Theor. Appl. Genet.,97: 1248–1255.
RAY-CHOUDHURY, P., S. KOHLI, K. SRINIVASAN, T. MOHAPATRA and R.P. SHARMA (2001): Identification and classification of aromatic rice based on DNA fingerprinting. Euphytica, 118: 243–251.
ROHLF, F.J. (2000): NTSYS-pc version 1.70 numerical taxonomy and multivariate analysis system. - Applied Biostatistics Inc., Exeter Software. Setauket - New York.
THOMPSON, J.A., R.L. ELSON and L.O. VODKIN (1998): Identification of diverse soybean germplasm using RAPD markers. Crop Sci., 38: 1348–1355.
VIRK, P.S., B.V. FORD-LLOYD, M.T. JACKSON and H.J. NEWBURY (1995): Use of RAPD for the study of diversity within plant germplasm collections. Heredity, 4: 170–179.
WILLIAMS, J.G.K., A.R. KUBELIK, K.J. LIVAK, J.A. RAFALSKI and S.V. TINJOY (1990): DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucl. Acid Res., 18: 6531-6535.
ISPITIVANJE EFIKASNOSTI RAPD MARKERA U RAZVOJU MOLEKULARNOG PROFILA ZA ISPITIVANJE GENETI KE ISTO E (Glycine max L.)
Sanjay KUMAR
Geneti ka laboratorija, Odelenje za zoologiju Banaras Hindu Univerzitet, Varanasi, Indija
Izvod
Glavni napredak u istraživanjima soje je u razumevanju geneti kih osobina i primena novih tehnologija u njenom poboljšanju. Utvr ivanje geneti kog odnosa aktivnosti enzima i molekularnih markera je pokazalo da je konzistentno sa o ekivanjima zasnovanim na poreklu i informacijama pedigrea. Da bi se identifikovali efikasni markeri koji e biti koriš eni u ispitivanjima geneti ke isto e polimorfizam je osnovni kriterijum. U ovom radu vršena je evaluacije 80 RAPD markera od kojih je moglo da se amplifikuje 37 a samo 30 prajmera je dalo nedvosmislene profile. Od tih 30, 22 je imalo polimorfnu sliku fragmenata DNK (banding). Analiziranih 30 RAPD markera je dovelo do umnožavanja 120 fragmenata od kojih je 81 (67.5 %) fragmenata bilo polimorfno. U proseku dobijena su 4 fragmenta po prajmeru i 115 prajmera (50 %) je dalo ve i broj fragmenata od prose nog broja što je ohrabruju e. 8 prajmera je imalo 100% polimorfizam. Rezultati su indikativni za efikasnost RAPD prajmera u razvoju molekularnih profila.