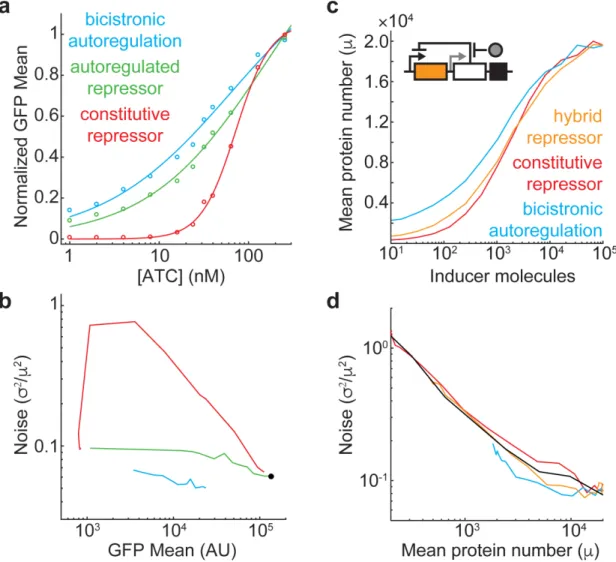
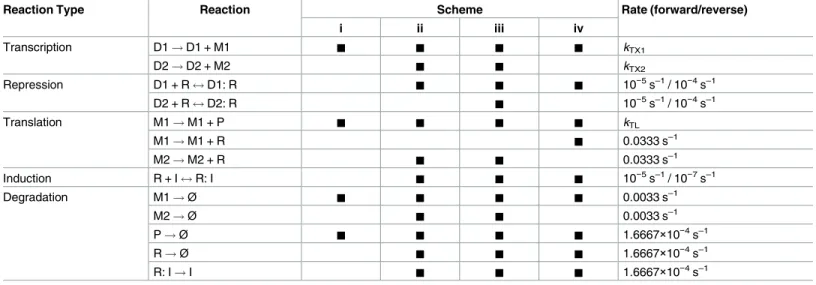
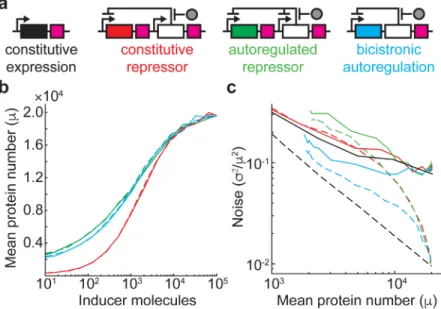
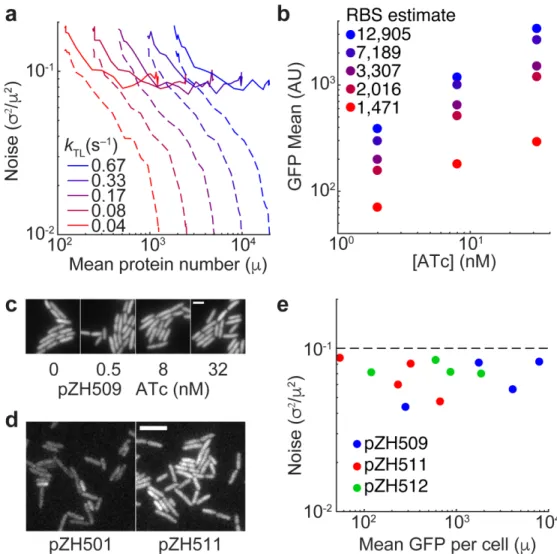
A plasmid-based Escherichia coli gene expression system with cell-to-cell variation below the extrinsic noise limit
Texto
Imagem

Documentos relacionados
The alkane hydroxylase ( alkB ) gene of Pseudomonas putida GPo1 was constructed in a pCom8 expression vector, and the pCom8-GPo1 alkB plasmid was transformed into Escherichia coli
An expression plasmid (pCFA-1) carrying the cfaB gene that codes for the enterotoxigenic Escherichia coli (ETEC) fimbrial adhesin coloni- zation factor antigen I (CFA/I) subunit
Considering the region as a whole, in the Latin American Political Science Asso- ciation (ALACIP) congress that took place in Buenos Aires in July 2010 just eight papers of 1,230
Understanding the organization and function of transcriptional regulatory networks by analyzing high-throughput gene expression profiles is a key problem in computational biology.
In one such case involving the endogenous immunoglobulin heavy chain (IgH) locus, removal of the enhancer from the IgH locus of a B cell line creates conditions in which the heavy
agreement with the SMN2 gene copy number and mRNA expression levels, Western blot analysis with an antibody that specifically detects human SMN revealed that in the absence
We found that the nongenetic (cellular) memory of yEGFP::zeoR expression and individual cell division rates at specific gene expression states were necessary to predict overall
Figure 5 - qRT-PCR: T0 represents the negative control for AFMSC gene expression; A1: Newly differentiated cell gene expression; chondro represents adult human articular chondrocytes