Vol-7, Special Issue-Number4-June, 2016, pp700-712 http://www.bipublication.com
Research Article
Investigating the inhibitory effect of bismuth nanoparticles on E. coli (with
pks gene) extracted from the cancer tumors of colorectal
Sadigheh Mehrabiyan* and Roghaye Kiyani School of Biological Sciences,
Islamic Azad University, Tehran-North Branch, Tehran, Iran
ABSTRACT
Increasing antibiotic resistance in the community necessitates the need to produce and develop new antimicrobial substances. Today the use of metal nanoparticles as antimicrobial materials is highly regarded. In this study, antimicrobial effects of bismuth nanoparticles on E.coli strains with pks gene – producers of colibaktin protein, predisposing factor for colorectal cancer were studied. After identifying strains containing the gene by PCR, different antimicrobial tests were conducted on the species. E.coli strains to tetracycline antibiotics, metronidazole, ciprofloxacin, and norfloxacin were sensitive or semi-sensitive and bismuth nanoparticles not had antimicrobial effect on strains tested. However, the combination of bismuth nanoparticles and each of antibiotics was as variable between species with synergistic effect. Regarding the existence of synergism between antibiotics used and bismuth nanoparticles, to treat intestinal infections can be used lower doses of antibiotics or fewer types of antibiotics. Considering the carcinogenic of strains, susceptibility to colorectal cancer can be prevented.
Keywords: bismuth nanoparticles, colorectal cancer, Escherichia coli, pks gene, Colibaktin
INTRODUCTION
Since the identification of bacteria, human beings have always been seeking to find an effective drug against infections caused by them and bacteria also achieved to effective mechanisms to eliminate antibiotics. Today, with the emergence of drug resistance among pathogenic bacteria, treatment of infectious diseases is faced with many problems. By the development of resistant organisms to antibiotic compounds, today we observe numerous reports on their widespread outbreaks in various parts of hospitals that often are the result of broad-spectrum beta-lactam drugs (Kollef et al, 2001). One of the most important factors for colorectal cancer is bacterial infections. Surveys show that infection with certain types of bacteria of Escherichia coli (E.coli) in patients with
inflammatory bowel diseases, particularly
ulcerative colitis can cause colorectal cancer. These bacteria using existing inflammatory provide conditions of the intestinal flora for their pathogenesis. The bacteria by producing toxins
secreted of Colibaktin that is as secondary metabolites of bacteria, with disruption in cell
cycle causes initiation, progression, and
development of colorectal cancer (Dastjany F. et al., 2015).
This bacterium also is the most important causes of microbial in urinary infections and is considered the agent of many hospital infections such as sepsis, wound infections, gastroenteritis and neonatal meningitis. E. coli is one of the opportunistic pathogens for hospital and due to the acquisition of plasmids that are broad-spectrum beta-lactamase encoding, are resistant to beta-lactam antibiotics. For this reason, treatment of infections caused by E. coli is faced with difficulty (Francesco, 2007). ). Some strains of E.coli bacteria has a specific genomic region called pks that this area produces peptide polyketide synthetase that leads to the
production of toxic called Colibaktin
seen in the frequency of mutations in their genes. Studies have shown that infection can be caused mutagenic potential. So, colonization of strains of E.coli bacteria with pks gene can contribute to the development of colon cancer (Parkin et al, 2007). Today, nanotechnology is used significantly in the development and introduction of antimicrobial and antibacterial effective in the treatment of some incurable diseases such as cancer. It is well known that nanoparticles compared to natural antibiotics as well as synthetic for antimicrobial activities have some of the considerable advantages. For example, the Staphylococcus aureus is a bacterium that is naturally sensitive to all antibiotics. Among the various nanoparticles, metal nanoparticles have special features according to its unique features that exactly are in relation to their nanometer dimensions, and never have in mass state such potential behavior. Clearly, only a very small amount of the metal ions (such as copper, iron, cobalt) are required for both eukaryotic and prokaryotic cells. For example, iron is a cofactor for many enzymes. It also plays a vital role in many physiological processes such as DNA replication, transcription and metabolism (Andreini et al, 2008). Among these nanoparticles, silver, copper, gold, aluminum, titanium, zinc and recently cerium and Bisut are also applied in this area.
Resistance, often can be obtained through horizontal gene transfer from external sources and may be associated with their ability to produce beta-lactamase enzymes (neutralizing penny Celine) and break down beta-lactam ring linked by the enzyme (Chambers and Deleo 2009, Seil and Websters 2012). On the other hand, the mechanisms of antimicrobial of nanoparticles are likely linked to surface area of the particles, morphology of unusual crystal (edges and corners), and reactive sites. In other words, the smallest particles have the strongest antimicrobial effect (Seil and Websters 2012). So, very likely size will have a decisive role in the bactericidal properties of nanoparticles. Thus, the use of broad-spectrum antimicrobial material to eliminate resistant bacteria of intestine can be effective in preventing cancer. This study investigates the antimicrobial effect
of solution of bismuth nanoparticles on E.coli strains carrying the pks genes as well as antimicrobial effects of nanoparticles with several common antibiotics in the treatment of intestinal infection are compared.
MATERIALS AND METHODS Materials used
Bismuth nanoparticles (BNPs) (a spherical morphology with a particle size of 170-200nm, spherical with irregular peripheral and with early concentration of 3500 ppm) (Figure 1), sterile distilled water and physiology serum 0.9% sodium chloride, crystal violet, Safranin , alcohol - Estonia, cotton, foil, Para, antibiotics
tetracycline, metronidazole, ciprofloxacin,
norfloxacin in the form of powder from Merck companies of Germany, Sigma-Aldrich America and others were prepared in lab and were used without initial modification. Mueller Hinton Broth culture media (Liquid Microbial Culture) (LMC, LB) and Mueller-Hinton agar (Solid Microbial Culture) (SMC) were used for culturing bacteria. In addition, E.coli strains isolated from patients with colorectal cancer with pks genes, and standard strain of S.aureus ATCC 6537 was prepared ready.
Equipment needed
The equipment used in this project include: spectrophotometer, Incubator, autoclave, Vertex, oven, flame, pipette in volumes 1 and 10 mm, Pasteur pipette, long and short test tube, cylinders, Erlenmeyer flasks in different volumes , disposable plates, sampler, lam, Fildo platinum (loop), track, anise, dropper, optical microscopy and digital scales.
Preparing liquid and solid culture media required
To prepare 1 liter of SMC, weight 34 grams of it carefully and pour into a 2-liter flask and add 1000 ml of distilled water to it and close the flask door with cotton and foil. In the next stage autoclave the media and after cooling the media distribute them near the flame in plates of 10 and 8 cm. For preparing SMC slant before autoclaving put the flask on heaters until all ingredients are well resolved and the uniform medium is obtained. Then distribute the medium in long test tubes and cover tubes door with cork and foil, then placed in the autoclave. Then before cooling the inside medium of tubes, put them on a slope surface to media form as slant. Also, to prepare 200 ml of LB Liquid Broth, we weight the amount of 4.2 grams of the medium mentioned and transmit into bottle of 500 ml. Then, with a graduated cylinder, add 200 ml of distilled water and put on heater to media solved in distilled water and obtained an uniform medium, then with one ml pipette, transmit as much as 1 mL of this uniformed mixture into short tubes of test and like previous medium, we cover the tubes with cotton and foil, then autoclave the media and then are ready to use after cooling.
Isolation of strains containing pks gene using PCR
DNA extraction process
First take E.coli bacteria from single clone and grow in 5 ml LB liquid medium. We put 24 hours in incubation at 37 ° C with a rotation speed of 200 rpm. A bacterium grows. Then, pour 2 ml of the liquid medium into micro-tubes and centrifuge 10 minutes with rotation speed of 7500 rpm and discard the supernatant and on the sediment remaining pour 100 micro liters of buffer protease and 5 micro liters protease ensyme, then vortex and keep 30 min at 55 ° C living.
In addition, pour 400 micro-liters of liquid of lysis buffer and vertex for 15 to 30 seconds and then pour 30 micro-liters of precipitation solution and we vortex for 5 seconds. Then, we centrifuge for 10 minutes at 12000 rpm. We discard supernatant and add to the sediment remaining 1 ml washing buffer and vertex for 5
seconds. Then we centrifuge with12000 rpm for 5 minutes. Again discard supernatant buffer and put the remaining sediment at the bottom of the micro-tubes openly in the same state at a temperature of 65 ° C for 5 minutes. Then, add 50 micro-liters of buffer solvent to micro-tubes and then vertex, and in continue leave micro-tubes in the closed position for 5 to 10 minutes at a temperature of 65 ° C. Then we centrifuge 12000rpm for 30 seconds away. The supernatant contains DNA of bacteria Escherichia coli (E.coli). Finally, to evaluate the quality of extracted genomic DNA, sample obtained on agarose gel 0.8%, was electrophoresed. To evaluate the quantity of extracted genomic, electrophoresis was used. For this purpose, first extracted DNA solution was diluted with amount of 100 times and its light absorption was measured at a wavelength of 260 and 280 nm with a spectrophotometer. If the DNA absorption ratio in 260 to 280 is 1.8, DNA extracted is free from phenolic pollution, protein and RNA.
Determine minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) for BNPs
Before anything a suspension of considered bacteria should be prepared to be used in future studies. To do so, with the sterile loop on the flame, near the flame of Clonies of each microorganisms grown on the surface of slant culture, some should be taken and in aseptic conditions will be transmitted 0.9% sodium chloride to the long test tube containing 10 ml of physiology serum to be set the suspension turbidity obtained in the spectrophotometer at a wavelength of 600nm to 0.1 that the amount of light absorption is equivalent to 108 CFU / ml. Then, to determine MIC and MBC bismuth nanoparticles, the method presented in reference (Wei et al, 2009) was used as follows.
and as serial dilution, we go forward then to maintain balance, take 1 ml from tube 13 contents and remove. Repeat this process for each of the microorganisms. Then, 100 ml of microorganisms is added to each of the tubes from NO 1 to 12 as well as tube NO 14. Finally put the test tubes on vortex to medium, BNP and microorganisms that here is E.coli bacteria well mixed with each other, and then placed tubes for 24 hours in incubator at 37 ° C. After this time, the tubes should be investigated in terms of formation of turbidity. The lowest concentration where turbidity was not seen is considered as the minimum inhibitory concentration (BNP). For SMC (solid medium) (Mueller-Hinton agar) act respectively above and after 24 hours of incubation at 37 ° C, the plates in terms of growth of microorganisms will be checked on them, the lowest concentration where growth was not seen in them as the minimum bactericidal concentration (MBC) is considered. It is worth noting that, in the test, tube No. 13 is without inoculation (without microorganisms) and as a negative control, and tube NO.14
without BNP with inoculation of
microorganisms is considered as a positive control. BNP concentration range from 1750 ppm in the test tube of No. 1 to 0/85 ppm has been in tube NO 13. In addition, this method was applied to all microorganisms as mentioned respectively. It should be noted that solvent of all anti-veterinary used in this study has been according to the instructions provided by the manufacturer company of distilled water. To achieve the best results, different concentrations of nanoparticles and microorganisms were tested together to ultimately the best concentration for whichever that is considered the most effective.
Disk Diffusion Test
After preparation of the bacterial suspension with a concentration of 108 CFU / ml, sterile swab is dipped in suspension and transmitted on MHA culture and distributed on the entire surface of the plate. After surface media on the plate, using sterile Pasteur pipette, some wells will be created with correct distance on the plate and value of 30 ml of BNP solution in different concentrations is transferred in the wells. Then, the plates were incubated for 24 hours at 37 ° C.
It should be noted that the antibiotics used in this test is purchased as disk and antibiotics disks were placed on the plate. After incubation, the diameter of non-growth halo was measured and recorded on the plate. For each of the strains, test was administered three times and the average of diameter of halos was considered as the final result.
Kill Time Determination Test
First liquid medium of MHB (Mueller Hinton
Broth) with a known concentration of
nanoparticles and antibiotics is prepared, according to what mentioned in the method of determining MIC and MBC (concentration of MIC and a higher concentration and lower than it) and then from the bacterial suspension already prepared is added to the culture medium as after inoculation per 1 ml of medium, the number of 108 bacteria is existed. After a little insemination, environment is perfectly stirred and OD zero time t = 0 is read by spectrophotometer. After that, the medium is placed in shaker incubator in temperature of 37 ° C and the second time t = 2 h is read again. Thus, in the hours of 4, 6, and 8, OD is read. After each visit OD, the amount of 100 µl of MHB is removed and after appropriate dilution, on a 10cm plate containing solid culture medium (agar), MHA will be distributed. The number of colonies after 24 hours of incubation at 37 ° C is counted. Since, the test is repeated three time, the average of number of counted colonies per plate as the final number will be recorded. To evaluate the trend of increasing or reducing the bacterial population, logarithmic graph of the number of bacteria at different times (nt) in the number of initial bacteria at time zero (N0) in terms of time (t) (s) (log Nt / N0vs. T) for each the tested strains and antimicrobial substances is drawn.
It should be noted that, one of the culture medias with lack of BNPs or antibiotics where breeding is done along with other cultures as positive control can be incubated. Thus, we will have bacterial growth curve in normal conditions on the graph. The synergism effect in combination of bismuth nanoparticles and antibiotics tested
DISCUSSION AND CONCLUSION
Isolate E.coli strains carrying pks genes using PCR
Using primers introduced in the Materials and Methods, strains carrying pks genes were
isolated, figures 2 and 3. During the study, questionnaires from patients with CRC were prepared that the specifications of patients with strains E.coli carrying pks genes are listed in table 1.
Figure 2. Agarose gel image of PCR products
Table 1. Profile of patients with strains carrying the pks gene
Strain No. age gender race smoking Eating red meat Eating vegetables UC‒19 48 Female Fars No Three times a week Yes
UC‒21 60 Male Fars Yes Three times a week No
UC‒33 37 Female Kord No Once a week No
Disk diffusion test results
After measuring the non-growth halos of each of the antimicrobial agents used (antibiotics, BNP, (3500ppm), bismuth subcitrate) values obtained with the values in the table CLSI (Table 2) were compared (Clinical and Laboratory Standards Institute, 2014) and resistance pattern of each of the strains were determined, respectively (table 3).
Table 2. The diameter of non-growth halos and pattern of antibiotic resistance of each of the strains BSN (250 mg/ml) BNPS
(350ppm) Tet
Met NOR
Cip Strain No.
0
)R( 0
)R(
13 mm
)I(
14 mm
)S(
20mm
)S(
26 mm
)S( UC‒33
0
)R(
0
)R(
15 mm
)S(
12 mm
)S(
21mm
)S(
23 mm
)S( UC‒21
0
)R(
0
)R(
14 mm
)I(
15 mm
)S(
18 mm
)S(
24 mm
)S( UC‒19
R: Resistant, I: Intermediate, S: Sensitive, CIP: ciprofloxacin, NOR: norfloxacin, BSS: Bismuth subcitrate, BNPs: Bismuth nanoparticles, Tet: tetracycline
According to the results of Disk diffusion test, bacterial strains were sensitive to the used antibiotic bacteria than tetracycline that two of the strains were semi-sensitive. However bismuth subcitrate and bismuth nanoparticles not have any antimicrobial effect on the bacteria used.
Table 3. Table CLSI number MO7, and special MO2 Enterobacteriaceace
Interpretive criteria MIC (mg/ml) Diameter criteria(mm)
Zone interpretive Disk
content Antimicrobial
Agent
R I
S R
I S
8 12-14
Tetracycline
2 16-20
8 13-16 Norfloxacin 16 ‒ ‒ ‒ Metronidazole
R: Resistant, I: Intermediate, S: Sensitive,
Since the bismuth nanoparticles not had antimicrobial effect on E.coli gram-negative bacterium, its antimicrobial effect on gram-positive bacteria of S.aureus using Disk Diffusion were evaluated that the results obtained showed the sensitivity of the bacteria to BNPs.
Table 4. Pattern of antibiotic resistance of S.aureus ATCC6538
BSN BNPs Tet NOR Cip Microorganism 0(R) 13mm(S) 20mm(S) 22mm(S) 25mm(S) S.aureus
The values obtained were compared with CLSI table (Table 5), thus susceptibility of this strain were also examined.
Table 5. CLSI table number MO2, MO7 especially Staphylococcus:
Interpretive criteria
MIC Diameter criteria (mm)
Zone interpretive Disk content Antimicrobial Agent R I S R I S 8 15-18 Tet 2 16-20 CIP 8 13-16 NoR
Results of MIC, MBC of bismuth nanoparticles and used antibiotics
To determine the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC), diluting liquid medium (Broth Tube Macrodilution) was used. MBC and MIC values, for each of the antimicrobial substances used are in the table (6). Then, the resulting values with the values CLSI (Tables 2 and 5) were compared in terms of it, resistance patterns of each of the strains were determined (Table 6). Despite the lack of sensitivity of E.coli strains towards BNPs with concentration of 3500ppm, S.aureus of used standard was sensitive to the nanoparticles with concentration of 350ppm.
MIC and MBC results, combinations of each of the antibiotics with BNPs
After examining the values MIC and MBC (µg / ml), since BNPs studied had no effect of anti-microbial on strains of E.coli as well as the non-growth halo related to BNPs was zero, BNPs synergistic effects with each of antibiotics have been studied and the results were collected in table 6.
Table 6. Quantities of MIC, MBC (µg / mL) obtained for each of antimicrobials
BNPs Met Tet NOR CIP strain No. MBC MIC MBC MIC MBC MIC MBC MIC MBC MIC E.coli 0 0 16 8 16 8 6 3 2 1
UC‒33
0 0 16 8 8 4 8 4 2 1
UC‒21
0 0 16 8 16 8 6 3 2 1
UC‒19
350
µ
( g/ml) 350
)µg/ml(
‒ ‒ 8 4 6 3 2 1 S.aureus
Table 7. Antibiotic resistance patterns of bacterial strains tested (combination of antibiotics with BNPs) Met + BNPs
)µg/ml( Tet + BNPs
)µg/ml( NOR + BNPs
)µg/ml( Cip + BNPs
)µg/ml( Microorganism MBC MIC MBC MIC MBC MIC MBC MIC E.coli 16 8 8 4 3 1/5 2 1
U‒C33
6 3 16 4 4 2 1 0/5
U‒C21
12 6 8 4 6 3 1 0/5
U‒C19
‒ ‒ 6 3 4 2 1 0.5 S.aureus Time-Kill Test Results
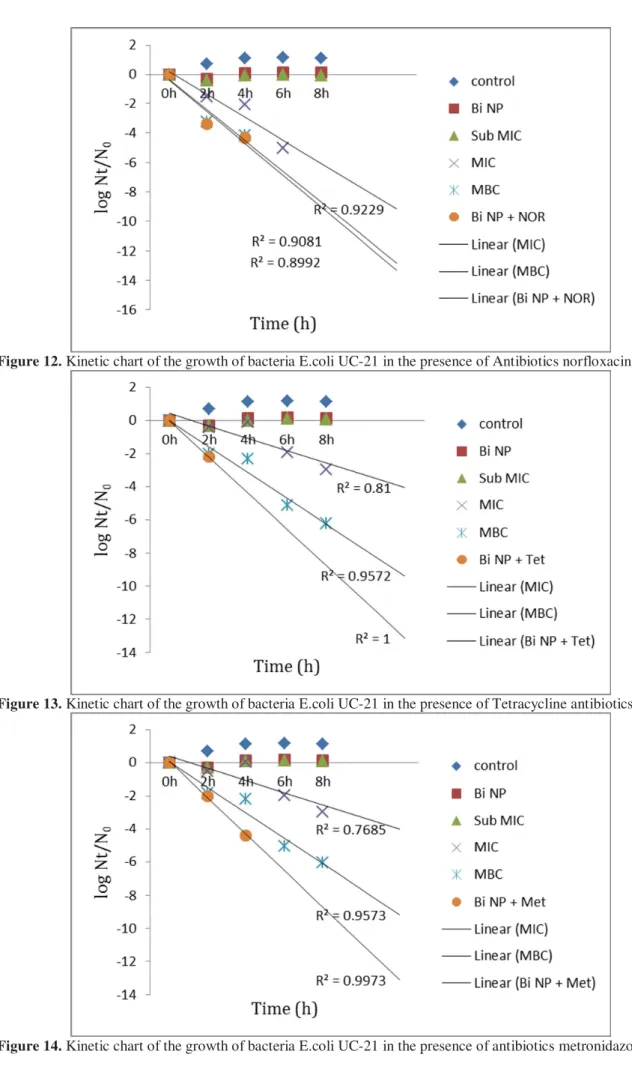
concentrations of antibiotics used, BNP [3500ppm], antibiotic compounds and BNPs can be studied (Charts of Figures 3 to 18).
Figure 3. Kinetic chart of the growth of bacteria E.coli UC-33 in the presence of antibiotics ciprofloxacin
Figure 4. Kinetic chart of the growth of bacteria E.coli UC-33 in the presence of antibiotics norfloxacin
Figure 6. Kinetic chart of the growth of bacteria E.coli UC-33 in the presence of antibiotics metronidazole
Figure 7. Kinetic chart of the growth of bacteria E.coli UC-19 in the presence of antibiotics ciprofloxacin
Figure 9. Kinetic chart of the growth of bacteria E.coli UC-19 in the presence of Tetracycline antibiotics
Figure 10. Kinetic chart of the growth of bacteria E.coli UC-19 in the presence of Antibiotics metronidazole
Figure 12. Kinetic chart of the growth of bacteria E.coli UC-21 in the presence of Antibiotics norfloxacin
Figure 13. Kinetic chart of the growth of bacteria E.coli UC-21 in the presence of Tetracycline antibiotics
Figure 15. Kinetic chart of the growth of bacteria S.aureus ATCC6538 in the presence of antibiotics ciprofloxacin
Figure 16. Kinetic chart of the growth of bacteria S.aureus ATCC6538 in the presence of antibiotics norfloxacin
Figure 17. Kinetics figure of bacterial growth S.aureus ATCC6538 in the presence of TCAB
Table 8. Investigating effect of the positive synergistic (+) in strains UC-21, UC-33, UC-19 in the presence of different antimicrobial
Synergic effect P-value
Under-test Antimicrobial Chemicals Strain No.
+ 0.011
BNPs + NOR C
UC‒21
0.006 BNPs 0.008 SubMIC 0.05> MIC 0.05> MBC + 0.011
BNPs + CIP C
0.006 BNPs 0.008 Sub MIC 0.05> MIC 0.05> MBC + 0.011
BNPs + Met C
0.006 BNPs 0.023 Sub MIC 0.068 MIC 0.041 MBC + 0.011
BNPs + Tet C
0.006 BNPs 0.006 SubMIC 0.032 MIC 0.037 MBC + 0.011
BNPs + Tet C
UC‒33
0.006 BNPs 0.013 SubMIC 0.23 MIC 0.103 MBC + 0.011
BNPs + NOR C
0.006 BNPs 0.099 SubMIC 0.015 MIC 0.016 MBC + 0.011
BNPs + CIP C
UC‒19
0.006 BNPs 0.013 SubMIC 0.023 MI C 0.066 MBC + 0.011
BNPs + Tet C
0.006 BNPs 0.035 SubMIC 0.01 MIC 0.052 MBC + 0.011
BNPs + Met C
0.017 BNPs 0.017 SubMIC 0.018 MIC 0.106 MBC CONCLUSION
Inorganic Nanostructures have many
applications in life sciences and medical fields. Nanoparticles have had application as a coating for materials, treatment, and diagnosis of diseases (Colvin, 2003). Nanoparticles of titanium, silver, diamonds, iron oxides, carbon nanotubes and biodegradable polymers for use in diagnosis and treatment have been studied
that reduce the risk of spreading antimicrobial resistance (Hernandez et al, 2013).
In the present study, bismuth nanoparticles (BNPs) alone had antimicrobial effect on E.coli strains carrying pks gene.. However, in combination with antibiotics Met, Tet, NoR, and CIP exhibit significant antibacterial effect which indicates the synergistic antimicrobial effects doubled BNPs in the presence of these antibiotics. Patterns of synergistic effect in each of the strains are different. Synergistic effect occurs as increasing speed of the bacteria death as well as reducing the amount of MIC (minimum inhibitory concentration) of tested antibiotics.
However, regardless of the lack of bismuth nanoparticles antimicrobial effect on gram-negative bacteria E.coli, in the conducted study alone on the S.aureus bacteria had antimicrobial effect, as well as showed a significant synergistic effect in combination with tested antibiotics. Before by other researchers, the inhibitory effect of Bismuth nanoparticles on bacterial growth of Gram-positive Streptococcus mutans and biofilm formation of bacteria is shown (Hernandez et al, 2012) that the results are quite consistent with the results reported in the present study.
Thus, due to the low toxicity of bismuth metal (Bismuth, Bi) than other heavy metals, from BNPs can be used to reduce the dosage of antibiotics used for enteral infections. Thus, in addition to reducing the concentration of antibiotics consumed, we gain to less toxic therapy and reducing the spread of antibiotic resistance. In addition, due to more sensitiveness of bacteria of Gram-positive S.aureus than BNPs can be used to treat other diseases caused by Gram-positive bacteria.
SOURCES AND REFERENCES
1. Andreini C,Bertini I,Cavallaro G,Holliday GL,Thornton JM. 2008. Metalions in biological catalysis: from enzyme databases to general principles. J Biol In org chem.13:12051218.
2. Chambers HF, Deleo FR. 2009. Waves of Resistance: staphylococcus anreus in the
antibiotics Era. Nat Rev Microbiol7: 629641.
3. Clinical and Laboratory standards Institute. 2014. Table 2A Entero bacteriaceae, Mo2 and Mo7. 34910: 5058.
4. Colvin VL. 2003. The potential environmental impact of engineered nanomaterials. Nat Biotechnol: 21(10): 11661170.
5. Francesco MA, Giuseppe R, Laura P, Riccardo N, Nin M. 2007. Urinary tract infections in Brescia, Italy: Etiology of uropathogens and antimicrobial resistance off common Uropathogens. Med Sci Moni., 13(6):136144.
6. Hernandez–Delgadino R, VelascoArias D, MartinezSanmiguel JJ, et al. 2013. Bismuth oxide an aqueous Colloidal nanoparticles inhibit Candida albicans growth and biofilm formation. Int J Nanomedicine. 8:164552. 7. Hernandez–Delgadino R, Velasco–Arias D,
Diaz D, et al .2012. Zerovalent bismuth nanoparticles inhibit Streptococcus mutans growth and formation of biofilm .Int J Nanomedicine. 7: 210913.
8. Kollef MH, Fraser VJ. 2001. Antibiotic resistance in the intensive care unit. Ann Intern Med., 134 (4):298314.
9. Nougayrede JP , Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. 2006. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science, 313: 848851[PMID: 16902142] 10.Parkin C, Potter DM, Saunders WS. 2007.
DNA repair pathways involved in anaphase bridge formation. Genes, chromosomes and Cancer. 46: 52231.
11.Seil TS, Websters TJ. 2012. Antimicrobial applications of nanotechnology: methods and literature. Inter J Nanomed.7: 27672781. 12.Wei SW, Qian W, Ye Y, Ma X .2009. The
synthesis of chitosan-based silver Nanoparticles and their antibacterial activity .Carbohydrate Research. 344: 237582.
13.Dastjany Farahani F, Mohammad Ganji Sh,
Sohrabi M. 2015- Journal of Medical Microbiology of Iran. 9 (2): 26-31.