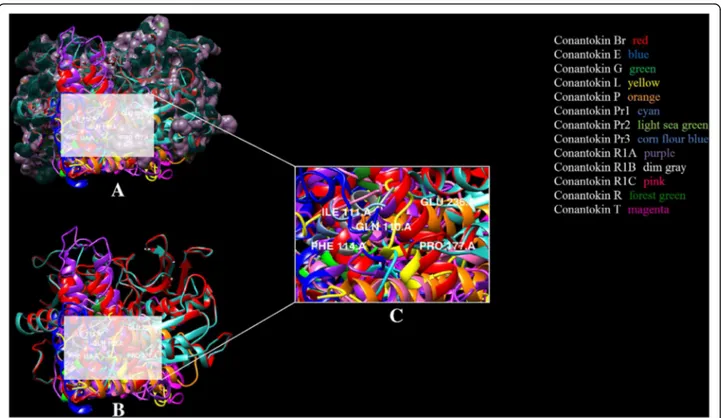
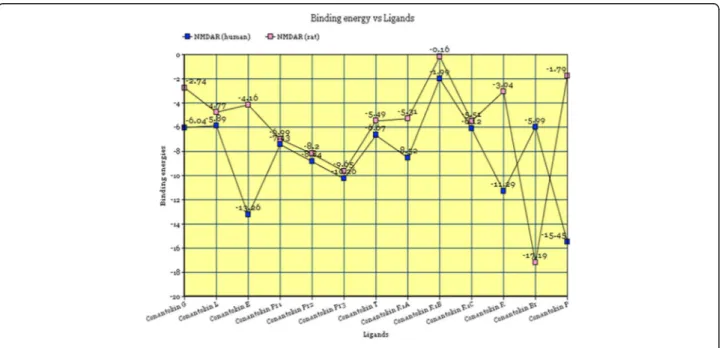
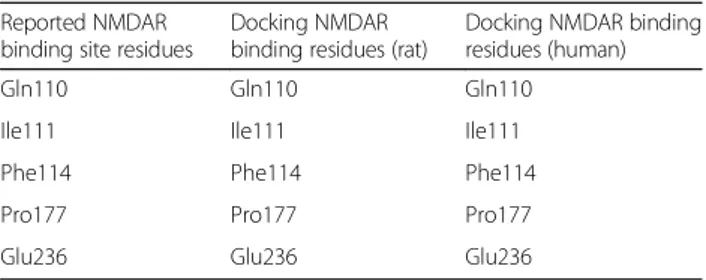
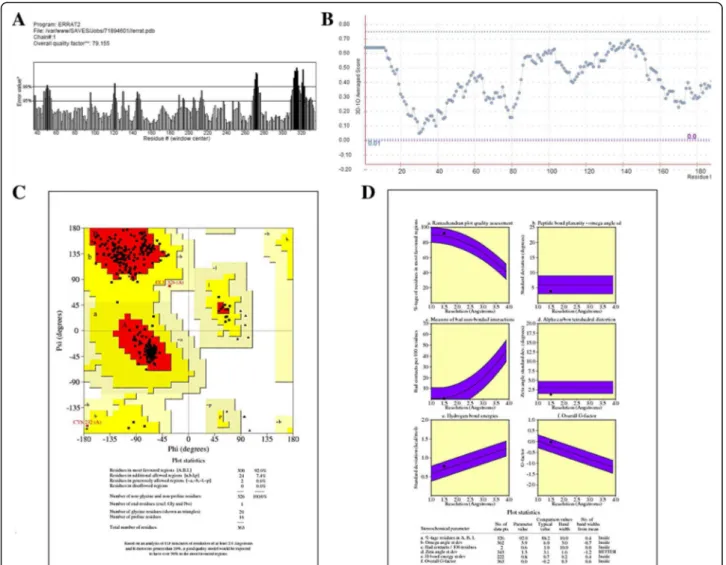
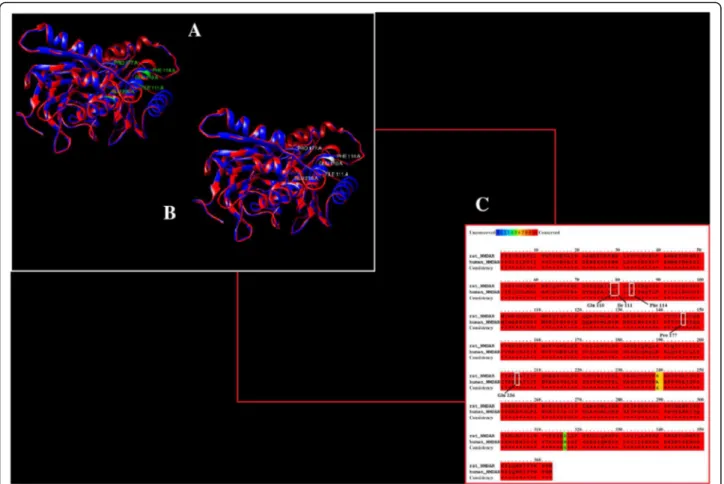
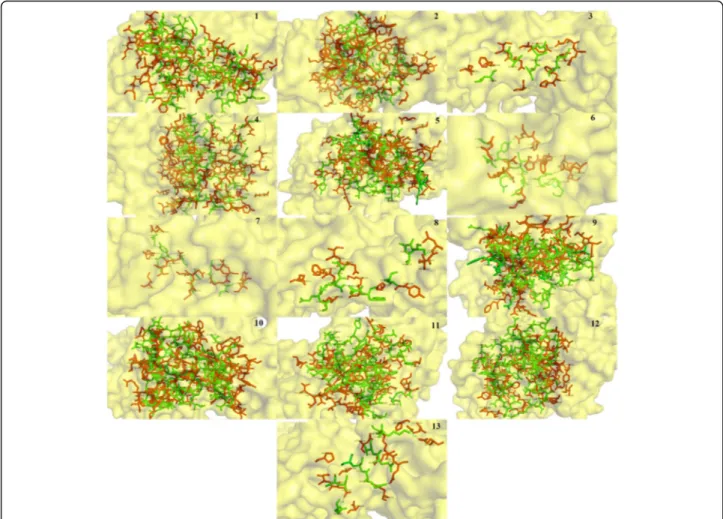
In silico analysis of binding interaction of conantokins with NMDA receptors for potential therapeutic use in Alzheimer ’s disease
Texto
Imagem

Documentos relacionados
Thus, an increase in the number of nAChRs composed of a 7 subunits (low affinity binding site) without changing the number of a 3ß4 subtype (high affinity binding site) could
The stare decisis doctrine in the United States legal culture requires that oncean appellate court in the State or federal judicial system has selected a principle
In addition to the amino acid residues forming the monosac- charide-binding site, other neighbouring residues located at the surface of the lectin also participate in the binding
Finally, we used alignments produced by our program to study binding site conservation in genome-wide binding data of key transcription factors in the Drosophila blastoderm, with
The number of binding sites and binding constant for naringin palmitate-BSA were less than in naringin-BSA, suggesting that the binding ability of naringin palmitate to BSA was
In this work, the feasibility of using salmon milt for REE recovery and separation was examined, along with the identification of the binding site of REEs in salmon milt..
At the first stage of the measurements results analysis, the gear wheel cast surface image was compared with the casting mould 3D-CAD model (fig.. Next, the measurements results
FIG. A) Close-in view of the arabinose binding site on the C-terminal domain of AraR, highlighting in blue shades the residues mutated by site-directed mutagenesis using the model