UNIVERSIDADE FEDERAL DO CEARÁ
FACULDADE DE FARMÁCIA, ODONTOLOGIA E ENFERMAGEM PROGRAMA DE PÓS-GRADUAÇÃO EM ODONTOLOGIA
DANIELA DA SILVA BEZERRA
ANÁLISE DA VIABILIDADE E EXPRESSÃO DE GENES DE VIRULÊNCIA DE
Streptococcus mutans EM LESÕES DENTINÁRIAS DE CRIANÇAS COM CÁRIE
PRECOCE DA INFÂNCIA
DANIELA DA SILVA BEZERRA
ANÁLISE DAVIABILIDADE E EXPRESSÃO DE GENES DE VIRULÊNCIA DE
Streptococcus mutans EM LESÕES DENTINÁRIAS DE CRIANÇAS COM CÁRIE
PRECOCE DA INFÂNCIA
Tese apresentada ao Programa de Pós-Graduação em Odontologia da Faculdade de Farmácia, Odontologia e Enfermagem da Universidade Federal do Ceará como um dos requisitos para a obtenção do Título de Doutora em Odontologia.
Área de concentração: Clínica Odontológica
Orientadora: Profa. Dra. Lidiany Karla Azevedo Rodrigues
Co-orientador: Prof. Dr. Rafael Nóbrega Stipp
Dados Internacionais de Catalogação na Publicação Universidade Federal do Ceará
Biblioteca de Ciências da Saúde
B469a Bezerra, Daniela da Silva.
Análise da viabilidade e expressão de genes de virulência de Streptococcus mutans em lesões dentinárias de crianças com cárie precoce da infância/ Daniela da Silva Bezerra. – 2014.
72 f. : il.
Tese (Doutorado) – Universidade Federal do Ceará, Faculdade de Farmácia, Odontologia e Enfermagem, Departamento de Clínica Odontológica, Programa de Pós-Graduação em Odontologia, Doutorado em Odontologia, Fortaleza, 2014.
Área de Concentração: Clínica Odontológica.
Orientação: Profa. Dra. Lidiany Karla Azevedo Rodrigues. Coorientação: Prof. Dr. Rafael Nóbrega Stipp.
1. Cárie Dentária. 2. Dentina. 3. RNA Mensageiro. 4. Reação em Cadeia da Polimerase em Tempo Real. 5. Expressão Gênica. I. Título.
A Deus, amado e venerado Pai, a quem eu entrego cegamente meus passos e minha vida.
Aos meus amados pais, Maria Nilsa da Silva Bezerra e Francismar Bezerra dos Santos. Obrigada pela vida, educação, dedicação e amor desmedidos e pela transformação do meu
mundo, mesmo nos momentos em que eu não percebia.
AGRADECIMENTOS ESPECIAIS
À minha orientadora, Profa. Dra. Lidiany Karla Azevedo Rodrigues, pela dedicação, competência e amizade ao longo dos anos de meu mestrado e doutorado. Por ser um exemplo de pessoa, pesquisadora e professora a serem seguidos.
Ao meu co-orientador, Prof. Dr. Rafael Nóbrega Stipp, pelo acolhimento, dedicação e ensinamentos valiosos.
Às companheiras de trabalho, Beatriz Gonçalves Neves e Sarah Florindo de Figueiredo Guedes, meus alicerces nesta pesquisa, amigas sempre tão dedicadas e prestativas, principalmente nos momentos mais difíceis. Muito obrigada por tudo!
AGRADECIMENTOS
À Universidade Federal do Ceará (UFC) por meio do seu Magnífico Reitor Prof. Dr. Jesualdo Pereira Farias.
À Faculdade de Farmácia, Odontologia e Enfermagem da Universidade Federal do Ceará na pessoa de sua diretora Profa. Dra. Maria Goretti Rodrigues de Queiroz e de seu vice-diretor Prof. Dr. Sérgio Lima Santiago.
À coordenação do Programa de Pós-graduação em Odontologia da UFC, por meio da Coordenadora Profa. Dra. Lidiany Azevedo Rodrigues e da vice-coordenadora Profa. Dra. Cristiane Sá Roriz Fonteles.
Ao coordenador do curso de Odontologia, Prof. Dr. Fabrício Bitu Sousa.
À Faculdade de Odontologia da Universidade de Pelotas (UFPel), na pessoa do Prof. Dr. Flávio Demarco, pela receptividade, ensinamentos e apoio durante meu doutorado sanduíche na instituição.
À Faculdade de Odontologia de Piracicaba, da Universidade de Campinas (UNICAMP), na pessoa do Prof. Dr. Rafael Nóbrega Stipp, pela acolhida e disponibilidade para realização de parte dos experimentos desta tese.
À Universidade Estadual do Ceará (UECE), por meio da Profa. Dra. Maria Izabel Florindo Guedes, pela receptividade, disponibilidade e ensinamentos para realização de parte desta pesquisa.
Aos professores da Pós-Graduação deste curso, pelos ensinamentos e experiências trocadas.
Aos funcionários do Curso de Odontologia, geralmente tão dispostos a ajudar e a solucionar problemas.
Ao técnico de laboratório David Queiroz pelo apoio, coleguismo e disponibilidade durante a pesquisa.
Às estimadas crianças e seus responsáveis que participaram desta pesquisa, aos diretores, professores e demais funcionários das escolas públicas municipais de Fortaleza (CE) que foram visitadas para seleção dos voluntários do estudo. Sem os mesmos este trabalho não teria sido realizado!
Aos alunos de iniciação científica, Elano Barbosa, Daniel Sampaio, Edyr Pereira e Caroline Salema e todos os outros alunos que contribuíram para o bom andamento do trabalho.
Às colegas de turma Ana Patricia Cavalcante, Beatriz Neves, Denise Lins, Fabianni Apolônio e Ramile Lima pelo companheirismo e amizade.
Às colegas Patrícia Oliveira, Juliana Paiva, Mary Anne Mello, Vanara Florêncio pela ajuda e troca de conhecimentos durante os anos de doutorado.
Às colegas de pós-graduação, estudantes da UFPel, Mabel Succa Salas e Helena Schuch, pelo apoio e amizade durante minha estadia em Pelotas, RS.
Às colegas de pós-graduação Juliana Nunes Botelho e Erika Harth pela acolhida, amizade e troca de experiências durante a estadia na cidade de Piraciacaba, SP.
À aluna de doutorado Livia Marques e ao Prof. Dr. Sérgio Málaga, do laboratório de Biologia Molecular da UECE, pela ajuda e contribuição na realização de parte desta pesquisa.
Aos meus familiares, principalmente aos meus avós Adelaide Ferreira da Silva, Pedro Pereira da Silva, Francisca Pereira Dutra (in memoriam) e Valdemar Bezerra Sampaio (in memoriam) pela fé e amor sempre depositados e por me entenderem pelos vários momentos de ausência durante esta empreitada.
Aos meus estimados amigos, pela terna compreensão nas diversas ocasiões em que tive que ausentar-me para dedicação aos estudos. Pelo amor e carinho dedicados a mim.
A todos aqueles que de alguma forma contribuíram e torceram para a conclusão deste trabalho.
“Os que esperam no Senhor adquirirão sempre novas forças, tomarão asas como de águia, correrão e não fatigarão, andarão e não desfalecerão.”
RESUMO
A cárie precoce da infância (CPI) é caracterizada pela presença de uma ou mais lesões em superfície dentária de crianças menores de 6 anos de idade, sendo considerada um problema de saúde pública. Streptococcus mutans (SM) é considerado o principal agente etiológico da doença e possui vários atributos de virulência que permitem sua sobrevivência na cavidade oral e, portanto, sua seleção em situações de estresse nesse ambiente. Neste contexto, este trabalho teve por objetivos: (a) padronizar um método de extração e purificação de RNA total para viabilizar estudos de expressão gênica bacteriana de lesões de cárie dentinária in vivo
(capítulo 1); (b) identificar e quantificar SMmetabolicamente ativo e analisar a expressão dos genes de virulência atpD, fabM, nox, pdhA e aguD em lesões dentinárias ativas e inativas (capítulo 2). Foi realizada a coleta de 29 amostras de dentina cariada ativa e de 16 amostras de lesões dentinárias inativas de crianças com CPI. As amostras foram submetidas à extração e purificação do RNA total por um método padronizado, com base no uso de lise mecânica e kit de extração comercial e, em seguida, submetidas às reações de transcrição reversa (RT) para obtenção do DNA complementar (cDNA). Com o intuito de comprovação da especificidade dos primers para os genes de virulência de SM em amostras in vivo, 06 amostras de cDNA foram amplificadas e submetidas ao sequenciamento de Sanger para cada primer. Após verificação da especificidade, reações em cadeia da polimerase quantitativa em tempo real da transcrição reversa (RT-qPCR) foram executadas para todas as amostras. Os resultados obtidos mostraram que o RNA bacteriano pôde ser extraído em adequada quantidade e qualidade pelo protocolo padronizado, viabilizando seu uso em estudos de expressão de genes bacterianos em dentina cariada (Capítulo 01). Os resultados da RT-qPCR mostraram que, apesar da baixa quantificação em relação aos estreptococos e bactérias totais, SM viáveis foram detectados com quantificação semelhante em lesões ativas e inativas (p>0.05). Apesar disso, maior expressão dos genes de virulência pdhA e aguD foi detectada nas lesões inativas (p≤0,05) (Capítulo 02). Concluiu-se que, após padronização de uma técnica eficaz de extração de RNA, reações de RT-qPCR foram executadas com eficiência para identificação, quantificação e expressão gênica de SM em amostras de dentina humana cariada. SM estavam viáveis nos dois grupos de lesões, porém só nas lesões dentinárias inativas mostraram maior expressão dos genes pdhA e aguD, provavelmente para viabilizar sua atividade metabólica nas condições menos favoráveis à sua sobrevivência, inerentes a este substrato.
ABSTRACT
Early childhood caries (ECC) is characterized by the presence of one or more lesions on dental surface of children under 6 years old, being considered a public health issue.
Streptococcus mutans (SM) is considered the main etiologic agent of the disease and has several virulence features that allow its survival in the oral cavity and, therefore, its selection under environmental stress situations. In this context, this research had as objectives; (a) to describe a method of total RNA extraction and purification to conduct studies on bacterial gene expression from in vivo dentine caries lesions (Chapter 01); (b) to identify and quantify the metabolically active SM and analyze the expression of its virulence genes atpD, fabM, nox, pdhA and aguD on active and arrested dentin lesions (Chapter 02). Twenty-nine samples of active dentin caries and 16 samples of arrested dentin lesions were collected from children with ECC. The samples were submitted to extraction and purification of total RNA through a customized method, based on the use of a mechanical lysis and a commercial extraction kit and submitted to reverse transcriptase (RT) reaction to obtain complementary DNA (cDNA). Next, to demonstrate the primers specificity for the virulence genes on in vivo samples, 06 cDNA samples were submitted to Sanger’s sequencing for each primer. After verifying the specificity, RT-qPCR (quantitative real-time reverse transcription polymerase chain reaction) were executed for all the samples. The results showed that the bacterial RNA could be extracted in appropriate quantity and quality through the customized protocol, making its use on gene expression studies on dentin affected by caries (Chapter 01). The RT-qPCR results showed that, despite the low quantification, if compared to total streptococci and total bacteria, SM was detected as metabolically active with similar quantification on active and arrested lesions (p>0.05). Despite that, a higher expression of the virulence genes pdhA and
aguD was detected on arrested lesions (p≤0,05) (Chapter 02). It was concluded that after standardization of an effective RNA extraction technique, reactions of RT-qPCR were efficiently executed for SM identification, quantification, and gene expression on samples of human dentin affected by caries. This bacterium proved to be viable, however, only inactive lesions showed larger expression of the genes pdhA and aguD, probably to make its metabolic activity viable, even in unfavorable survival conditions.
SUMÁRIO
RESUMO
ABSTRACT
1.INTRODUÇÃO GERAL 14
2.PROPOSIÇÃO 18
3.CAPÍTULOS 19
3.1. CAPITULO 1 20
Extraction and purification of total RNA from human carious dentine: an approach to enable bacterial gene expression studies 3.1.1 Introduction 22
3.1.2. Material and Method 23
3.1.3. Results 27
3.1.4. Discussion 27
3.1.5. References 29
3.2. CAPÍTULO 2 37
Virulence gene expression of viable Streptococcus mutans in active dentine lesions from children with early childhood caries 3.2.1 Introduction 39
3.2.2. Material and Method 40
3.2.3. Results 45
3.2.4. Discussion 45
3.2.5. References 49
4. CONCLUSÃO GERAL 62
REFERÊNCIAS GERAIS 63
APÊNDICES 68
1. INTRODUÇÃO GERAL
A cárie dentária é uma doença caracterizada pela desmineralização do esmalte e da dentina e é dependente da presença de biofilme microbiano e açúcares (FEJERSKOV, 2004). Está entre as doenças mais comuns na infância (JIANG et al., 2014; PETERSON et al., 2013) e quando ocorre em uma ou mais superfícies dentárias cavitadas ou não, perdidas ou restauradas em crianças menores de 71 meses de idade é denominada de cárie precoce da infância (CPI) (AAPD, 2008), a qual pode resultar em considerável estado de dor e em impacto negativo na qualidade de vida (FILSTRUP et al., 2003; FEITOSA et al., 2005; ABANTO et al., 2011). A CPI uma doença polarizada, com a maioria das lesões distribuídas em um pequeno grupo de crianças (MACEK et al., 2004; STRÖMBERGet al., 2012; NUNES
et al., 2014), sendo mais prevalente em crianças de baixa renda de países em desenvolvimento e em regiões menos favorecidas de países desenvolvidos (BECKER et al., 2002; NUNES et al., 2014). No Brasil, a CPI é considerada um problema de saúde pública, visto que ainda tem uma elevada prevalência aos 5 anos de idade (53,4%) (MINISTÉRIO DA SAÚDE DE BRASIL, 2010).
Fatores externos como condições socioeconômicas, nível de educação e acesso à assistência médica (HALLETT & O’ROURKE, 2003, 2006; OLIVEIRA et al., 2008) podem ter um papel importante no estabelecimento da CPI. Além disso, fatores locais como disponibilidade de açúcares, flúor e substâncias antimicrobianas na cavidade oral (TEN CATE et al., 1998; THYLSTRUP et al., 1994) podem estar relacionados à alteração dos padrões de desenvolvimento e ao estado de atividade das lesões (HIRSCH et al., 2012; PIOVESAN et al., 2013).
inativadas por influência de fatores que modificam o ambiente oral, como a disponibilidade de açúcares e flúor, os quais podem modificar o pH e as condições de colonização microbiana. A atividade pode ser determinada pelo critério de Nyvad, um dos primeiros índices descritos e validados, o qual é baseado no diagnostico táctil (rugosidade da lesão e sensação táctil mediante sondagem) e visual (coloração e brilho) da lesão. Este sistema permite a diferenciação entre lesões ativas e inativas, cavitadas ou não, onde cada superfície dentária é classificada por um índice (MACHIULSKINE et al., 1998; NYVAD, 1999).
Mesmo sendo uma doença influenciada por diversos elementos, sabe-se que a microbiota é essencial para o aparecimento da cárie. Com o advento dos estudos moleculares para identificação e quantificação microbiana, foi possível detectar uma maior diversidade de bactérias relacionada com a doença (BECKER et al., 2002). Embora diversas espécies bacterianas possam estar envolvidas, os estreptococos do grupo mutans são os mais consistentemente associados (TANZER et al., 2001; MATTOS-GRANER et al., 2014) e, desde seu descobrimento por Clarke em 1924, Streptococcus mutans (SM) se tornou o microrganismo cariogênico mais estudado (BANAS et al., 2004). É uma bactéria acidogênica e acidúrica e está intimamente relacionada à iniciação e progressão da doença em adultos, crianças e em cárie precoce da infância (MATTOS-GRANER et al., 1998, 2001; BECKER et al., 2002; NOBRE-DOS-SANTOS et al., 2002; VACHIRAROJPISAN et al., 2004; SAXENA et al., 2008; TAKAHASHI & NYVAD, 2008). Em estudos prévios foi relatado que a sua cariogenicidade depende de uma complexa interação com as defesas do hospedeiro e com a microbiota residente (SIMPSON & RUSSELL, 1998; WEN & BURNE, 2002; ZENG & BURNE, 2008), influenciando sua virulência que reside em três atributos essenciais: habilidade para formar biofilme, produção de grandes quantidades de ácidos orgânicos (acidogenicidade) e tolerância ao estresse ambiental, particularmente pH baixo (aciduricidade) (LAW et al., 2007; SHEN et al., 2004).
Apesar do reconhecimento da sua patogenicidade na cárie dentária, estudos moleculares têm indicado divergências acerca da identificação e quantificação do
Streptococcus mutans em lesões cariosas na dentina, com indicação de baixa contagem ou até de ausência do mesmo (CHHOUR et al., 2005; AAS et al., 2008; MANTZOURANI et al.,
níveis de detecção, já que fatores de virulência expressos por esta espécie mostraram alterar a estrutura do biofilme e promover mudanças ecológicas levando ao predomínio de uma microbiota acidogênica e acidúrica, independente de sua abundância (MATTOS-GRANER et al., 2014). Da mesma forma, a despeito da prevalência de SM em lesões de cárie dentinária, é importante conhecer a sua atividade funcional e virulência neste tecido, ou seja, no próprio sítio da doença (NYVAD et al., 2013).
Alguns mecanismos metabólicos do SM, onde proteínas estão envolvidas, são responsáveis direta ou indiretamente pela sua virulência, sobrevivência e tolerância a estresses ambientais, como pH ácido, presença de oxigênio e disponibilidade de nutrientes. O sistema transportador de membrana F1-F0 ATPase (codificada pelo operon atpCDGAHF) é responsável pelo transporte de prótons H+ para fora da célula mesmo em condições de baixo pH, além de promover a produção de ATP, fonte de energia para o crescimento e persistência de SM (LEMOS & BURNE, 2008). Este comportamento propicia, no interior da célula, um pH mais alcalino quando comparado ao pH extracelular, sendo um dos mecanismos críticos na sua sobrevivência em ambientes ácidos (BANAS et al., 2004).
A enzima NADH oxidase (gene nox), participando da via glicolítica, promove a redução do oxigênio dissolvido no ambiente durante o crescimento bacteriano e produção de ATP, contribuindo para a sobrevivência da bactéria. Já a trans-2-cis-3-decenoyl-ACP isomerase (gene fabM) é responsável pelo incremento de ácidos graxos monoinsaturados da membrana plasmática quando há queda do pH externo (FOZO & QUIVEY, 2004).
Outro importante sistema é o da agmatina deaminase (AgDS), o qual é codificado pelos genes do operon aguBDAC (GRISWOLD et al, 2004). A agmatina entra na célula via
aguD, localizado na membrana, carreando putrescina e amônia para o meio extracelular. Esse sistema também propicia a síntese de ATP, viabilizando o crescimento bacteriano (GRISWOLD et al., 2006). Há, ainda, proteínas mais expressas em situações de depleção de nutrientes no ambiente oral, como a piruvato-desidrogenase (codificada pelo gene pdhA), que atua como alternativa para a continuidade da via glicolítica quando há queda nos níveis de glicose, além de viabilizar a formação de ATP para ser usado como fonte de energia (MOYE
et al., 2014).
progressão das lesões dentinárias (JIANG et al., 2014). Métodos baseados na análise do RNA, como qPCR, microarrays ou mesmo os sequenciamentos seriam desejáveis para este fim. Para isto, é importante que se tenha métodos eficientes para a obtenção de RNA de boa qualidade destas amostras, visto que sem uma amostra de ácido nucléico adequada, estudos da transcrição gênica não podem ser bem executados (BUSTIN et al., 2005; NOLAN et al., 2006). Entretanto, não há disponibilidade de técnicas moleculares descritas que contemplem esta abordagem em amostras orais que são coletadas em reduzida quantidade, como o tecido dentinário cariado, especialmente aquele originado de dentes decíduos, o que deveria ser encorajado para a condução de futuros estudos (NYVAD et al., 2013).
2. PROPOSIÇÃO
Esta tese de doutorado será apresentada em capítulos, tendo como objetivos:
CAPÍTULO 1: Padronizar um método eficiente de extração de RNA total de amostras de dentina cariada humana que viabilize estudos de expressão gênica das bactérias presentes neste substrato.
3. CAPÍTULOS
Esta dissertação está baseada no Artigo 46 do Regimento Interno do Programa de Pós- Graduação em Odontologia da Universidade Federal do Ceará, que regulamenta o formato alternativo para dissertações de Mestrado e teses de Doutorado e permite a inserção de artigos científicos de autoria e co-autoria do candidato. Por se tratarem de pesquisas envolvendo seres humanos, ou parte deles, o projeto de pesquisa referente a este trabalho foi submetido à apreciação do Comitê de Ética em Pesquisa da Faculdade de Medicina da Universidade Federal do Ceará, tendo sido aprovado sob protocolo nº 548.405 (ANEXO). Assim sendo, esta tese de Doutorado é composta por dois capítulos que contém artigos que serão submetidos à publicação em revistas científicas, conforme descrito abaixo:
CAPÍTULO 1
“Extraction and purification of total RNA from human carious dentin: an approach to enable bacterial gene expression studies”
Daniela S. Bezerra, Rafael N. Stipp, Sarah F. F. Guedes, Beatriz G. Neves, Lidiany K. A. Rodrigues
Este artigo será submetido à publicação no periódico “Journal of Microbiological Methods”.
CAPÍTULO 2
“Virulence gene expression of viable Streptococcus mutans in active and arrested dentine lesions from children with early childhood caries”
CAPÍTULO 1
Extraction and purification of total RNA from human carious dentine: an approach to enable bacterial gene expression studies
Daniela S. Bezerraa, Rafael N. Stippb, Beatriz G. Nevesa, Sarah F. F. Guedesa, Lidiany K. A. Rodriguesa*
a
Post-graduation Program, Faculty of Pharmacy, Dentistry and Nursing, Federal University of Ceará, Rua Monsenhor Furtado - Rodolfo Teófilo, Zip code: 60430-355, Fortaleza, Ceará, Brazil.
b
Department of Microbiology and Immunology, Piracicaba Dental School, State University of Campinas, Av. Limeira, 901, Zip code: 13414-018 Piracicaba, SP, Brasil.
Address all correspondence to:
Lidiany K. A. Rodrigues, DDS, MSc, PhD, Associate professor
Postgraduate Program in Dentistry, Faculty of Pharmacy, Dentistry and Nursing Federal University of Ceará, Fortaleza, Ceará, Brazil.
Rua Monsenhor Furtado - S/N - Rodolfo Teófilo – Zip code: 60430-355 Phone- #+558533668410/ Fax- #+558533668232
Email: lidianykarla@yahoo.com
Conflict of interests
There are no potential conflicts of interest for any of the authors.
Este artigo está escrito de acordo com as normas de publicação do periódico Journal of Microbiological Methods.
ABSTRACT
RNA isolation from bacteria within dentine caries lesions could be difficult due to reduced amount of collectable biomass and high mRNA instability. Attempting to overcome this challenge we describe one protocol developed to extract and purify total RNA from dentine lesions. The method was based on mechanical cell lysis and use of a commercial kit with guanidine. Quantity and purity of extracted RNA were measured with a microvolume UV-VIS spectrophotometer, RNA integrity was assessed by standard denaturing agarose gel electrophoresis and images were captured under ultraviolet light with camera and analyzed. DNase treatment removed genomic DNA and an additional step of purification was carried out in silica spin column. Final yield (ng/μl) was 68.40 ± 24.12, absorbance ratio (A260/A280) higher than 2.0 and RNA integrity were obtained. The purified samples were reversely transcribed and the expression of atpD and fabM gene from Streptococcus mutans analyzed by quantitative real-time PCR. The results demonstrated that the extraction methodology developed produced high-quality RNA from dentine microbiota for transcriptional analyses.
1. Introduction
Dental caries is a microbial disease dependent from biofilm formation in the presence of sugar (Fejerskov et al., 2004). The disease initiates with demineralization of tooth enamel and can progress to cavitation, destroying enamel and reaching dentine and pulp tissues (Simón-Soro et al., 2013).
To have a better understanding about microbial roles in dental caries process, several molecular methods of investigation have been adopted in the last years. Among them, RNA-based analysis focus in detection of metabolically active microbial members and identification of genes expressed under different circumstances (Nyvad et al., 2013; Twin et al., 2013). The majority of RNA-based investigations of oral microbiota analyzed only planktonic cells (Xu et al., 2011), in vitro biofilms (Cury et al., 2007, 2008; Klein et al., 2010; Friaz-Lopez and Duran-Pinedo et al., 2012; Stipp et al., 2013), or in vivo biofilms from rodent animals (Falsetta et al., 2012; Klein et al., 2012; Falsetta et al., 2014). Until now, the first metatranscriptomic studies of human dental biofilm and dentine were carried out (Benítez-Páez et al., 2014; Peterson et al., 2014; Simón-Soro et al., 2014). However, little information is given about technical procedures, RNA yield, purity and integrity from these human samples studies. Besides, more attention has been requested about transcriptional studies in human dentinal caries, which can reflect the behavior of cariogenic bacteria in loco (Nyvad et al., 2013)
The success of any transcriptional analysis depends not only on the amount of RNA extracted, but mainly on RNA quality, because the purity and integrity of this molecule can impact the accuracy of techniques such as RT-qPCR and microarrays (Bustin et al., 2005; Nolan et al., 2006; França et al., 2012a. Therefore, an adequate absorbance ratio A260/A280>2.0 (Imbeaud et al., 2005) and detection of rRNA bands without smearing (Bustin et al., 2005; Imbeaud et al., 2005) should be observed for verifying RNA quality.
Some particular limiting factors of carious dentine can hamper the obtaining of adequate quantity and quality of RNA, such as low amount of carious biomass per sample (Friaz-Lópes et al., 2012) and low proportion of bacteria when compared to other in vivo
Selinger et al., 2003); (3) rigid and thick cell walls of Gram-positive (Ludwig and Schleifer, 2000). Besides, RNA concentration and quality can be definitely influenced by adequate and well-conducted extraction and purification techniques based on the nature of the samples (Junttila et al., 2009; Nour t al., 2010; Rump et al., 2010; França et al., 2012a).
In an effort to solve possible challenges in RNA extraction from carious dentine and to establish an approach to enable bacterial transcriptional studies, the objective of this research was to customize a bacterial RNA extraction and purification method from human carious dentine.
2. Material and methods
2.1 Study population and ethical statements
One hundred eighty-one children from 48 to 71-month-old were examined in three public schools in Fortaleza, Ceará, Brazil. Fourteen children were selected according to the inclusion criteria: healthy children that have not recently taken antibiotics (3 months prior to the study); children with at least one primary teeth with carious surface with large distinct cavity and visible dentine without pulpal disease (confirmed by clinical and radiographic examination); the parent or guardian was willing to consent to the child’s clinical examination and dentine sampling. The study design was explained to the child’s parent or guardian, from whom informed consent was obtained if they were disposed to participate with their child. The study design, protocol, and informed consent were approved by the Ethics Committee of the Federal University of Ceará, protocol number 548.405.
2.2 Sample Collection
One tooth selected from each child was cleaned with pumice and local anesthesia was delivered. Carious teeth were isolated with rubber dam disinfected with 2% chlorhexidine. After removal of biofilm over the lesions, carious dentine were hand excavated with individual, sterile and nuclease free spoon excavators (S.S. White-Duflex, Rio de Janeiro, RJ, Brazil). These instruments were submitted previously to autoclaving sterilization and heated at 200°C for 18 h to eliminate RNases (Geiger et al., 1995).
freezer until RNA extraction onset. Cavities were then restored with a suitable restorative material and others visits were scheduled for the conclusion of other possible oral treatments.
2.3 RNA extraction and purification
The dentine samples were thawed and centrifuged (11,000 × g/ 1 min/ 4°C). The
RNAlater TM solution was removed using an automatic pipettor without disturbing the pellet. The samples were carefully transferred to cryogenic tubes containing 0.16 g of 0.1 mm diameter zirconium beads (Stipp et al., 2013). The mechanical disruption of bacterial cells was made by Mini-Beadbeater (Biospec Products Inc., Bartlesville, OK, USA) at maximum power (2 cycles of 60 s with 1 min rest on ice). Then, 850 �l of RLT buffer (Qiagen, Valencia, CA, USA) with 10% of β-mercaptoethanol (β-ME) was added and the suspension homogenized by vortexing. After centrifugation (11,000 × g/ 2 min/ 4°C), aliquots of 350 �l
of the supernatant were transferred to microtubes with 250 �l of pure ethanol, and vortexed. Next, the solutions were transferred to silica spin columns from RNeasy MinikitTM (Qiagen, Valencia, CA, USA) and centrifuged (11,000 × g/ 30 sec/ 20°C) to allow RNA binding to the
silica. After discarded the flow-through, 700 µl of RW1 buffer was added, centrifuged (11,000 × g/ 30 s/ 20°C) and discarded. Then, 500 µl of RPE buffer was added, centrifuged
(11,000 × g/ 30 s/ 20°C) and discarded. This operation was repeated. After additional
centrifugation (11,000 × g/ 2 min/ 20°C) for removal of residual RPE, the column was
positioned in a new tube and 40 µl of RNase-free water was dispensed on the center of it. After centrifugation (11,000 × g / 1 min/ 20°C), the RNA through the column and was
collected and immediately stored at -80°C freezer.
RNA concentration (ng/ �l) and purity (absorbance ratio A260/A280) were verified by Nanodrop 2000c microvolume spectrophotometer (Thermo Scientific, Wilmington, DE) and the integrity was verified by the detection of the 16S and 23S bands of rRNA in 1.2% formaldehyde agarose gel stained with ethidium bromide. Digital images were captured under ultraviolet light with a camera and analyzed by Gel Logic 200 Imaging System software (Eastman Kodak Co, Rochester, NY, USA).
The extracted and purified RNA samples were treated with TurboTM DNAse kit (Applied Biosystems, Ambiom, Austin, TX, USA) to removal genomic DNA. Five µl of enzyme and 5 µl of buffer from kit were added to RNA samples and incubated at 37°C for 15 min. Then, an additional purification step was executed by addition of 300 µl of RLT and 250 µl of pure ethanol to the samples. This solution was vortexed and transferred to silica spin columns from RNeasy MinieluteTM CleanUp kit (Qiagen, Dus, Bundesland, Germany). The purification steps previously described, including addition of RW1 and RPE buffers were repeated. Next, to elute the ultra-purified RNA, 40 µl of RNase-free water were added in the center of the silica column and centrifuged (11,000 × g/ 1 min/ 4°C). Concentration and
integrity of the RNA were checked.
2.5 cDNA synthesis
The cDNA were synthesized from the RNA using iScriptTM cDNA Synthesis Kit (Bio-rad, Hercules, CA, USA). Reverse transcription reactions were prepared with 6 µl of 5x iScript reaction mix, 1 µl of iScript reverse transcriptase, 1 µg of purified RNA and RNAse-free water in sufficient amount for final volume of 30 µl. The prepared solution was homogenized by vortexing for 5 s and incubated at 25°C for 5 min, heating at 42°C for 2 h and incubated at 85°C for 5 min in a Veriti® Thermal Cycler (Applied Biosystems, Foster
City, CA, USA). To acquire negative control samples of cDNA, synthesis reactions were carried out in the absence of reverse transcriptase enzyme to identify any residual genomic DNA after RT-qPCR. The acquired cDNA samples were normalized with addition of nuclease free-water to obtaining a concentration of 10 ng/ µl.
2.6 Sanger sequencing
Six cDNA samples were submitted to conventional PCR with primers for genes from
Streptococcus mutans (SM) related to tolerance acid: atpD
(5’-TGTTGATGGTCTGGGTGAAA-3’ and 5’- TTTGACGGTCTCCGATAACC-3’) (Xu et al.,
2011) and fabM (5’-CTGATTAATGCCAATGGGAAAGTC and
Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA, USA) and the reaction mix included 1 µl of Buffer PCR 10x, 1 µl of dNTPs 10mM, 2.5 µl of MgCl2, 3 µl of each primer F/R 10 µM, 0.25 µl of Taq polymerase 5 U/µl, 50 mM 1 µl of template cDNA (10 ng/µl) and 37.25 µl of nuclease-free water. The thermal-cycling consisted in 5 min at 95°C; 35 cycles of 15 s at 95°C, 60 s at melt temperature of each primer (table 2), 20 s at 72°C; 4 min at 60°C). Their products were purified with commercial kit (QIAquick® PCR Purification Kit, QUIAGEN, USA) and the sequencing reactions were performed using Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, USA) and submitted to thermal-cycling (20 s at 95°C; 40 cycles of 15 s at 96°C, 15 s at 50° C and 4 min at 60°C; 60s at 60°C). At the end, the products were precipitated, dried, resuspended in 10 µl of formamide and denatured for 5 min at 95°C. Sanger sequencing occurred in a 3500 Series Genetic Analyzer (8-capillary) (Applied Biosystems, Foster City, CA, USA). The sequencing data obtained was submitted to BioEdit software, version 7.2.5, to obtain contigs sequences, which were submitted to analysis using BLAST program (Basic Local Alignment Search Tool), available at http://estexplorer.biolinfo.org/hsd, to verify sequences similarity.
2.7 Reverse Transcription – Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
In order to get DNA from S. mutans UA159 for obtain standards curves, this strain was cultured in broth for 24 h as recommended by Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994). After centrifugation and washing in sterile saline solution (sodium chloride 0.9%), the quality and purity of bacterial cultures were checked by Gram staining and their DNA was recovered using an organic extraction protocol based on phenol/chloroform purification and alcohol precipitation (Wilson, 2001). Serial dilutions starting from 600 ng to 0.0003 ng (10-fold) of S. mtans DNA concentrations were used as standards and positive controls for relative quantification of the targeted bacteria. A standard amplification curve and a melting-point product curve were obtained for each primer set.
primer F/R 10 µM and 2 µl of cDNA (20 ng) was added in each spot of a 48-well plate. Assays were carried out in duplicate, and the final analyses were based on the mean of the two reactions. Negative control included reactions without template. The standard curves were used to transform the quantification cycle (Cq) values to the mass of cDNA amplified. The normalization of gene expression was made using the cDNA amount initially inserted in each spot of 48 well-plate (20 ng).
3. Results
Sample collected yielded 25.5 mg average of dentine. After applying the customized method of total RNA extraction and purification from this substrate, we obtained adequate RNA yield (67.01 ± 22.33 ng/ µl), purity (A260/ A280 = 2.00 ± 0.07) and integrity (table 02 and Fig. 01). Results of atpD and fabM gene expression for Streptococcus mutans were also shown in table 02. Amplification plot, standard curve and melt curve for atpD and fabM gene expression were presented in Fig. 02.
4. Discussion
Considering the importance of a better understanding of the bacterial behavior in carious sites (Nyvad et al., 2013), the current research established a methodology for RNA extraction to enable bacterial gene expression studies from human carious dentine samples. Until now, the majority transcriptional analysis of cariogenic bacteria were executed in planktonic cells in vitro and in vivo biofilms (Klein et al., 2010; Friaz-López et al., 2012; Falsetta et al., 2012; Peterson et al., 2014), substrates where higher bacterial concentration is found when compared to dentine lesions (Simón-Soro et al., 2013).
Low amount of sample and RNA instability are important limitations to study gene expression (Imbeaud et al., 2005; Raeymaekers, 1993) in in vivo oral samples (Friaz-López and Duran-Pinedo, 2012). In this study, the samples were collected from deciduous teeth and yielded an average weight of 25.5 mg of dentine per lesion, which is considered a low amount, when compared to other substrates, such as biofilms from in vitro studies (≅100 mg
showed yield (67.01 ± 22.33 ng/ µl), purity (A260/ A280 = 2.00 ± 0.07) and integrity of RNA sufficient for RT-qPCR assays.
Our adequate results were acquired from a customized method based on bead-beating and a commercial kit. Cell mechanical disruption has several advantages over the traditional methods of cell lysis (lysis buffer, manual disruption and liquid nitrogen) including reduction of work time, multiple sample extraction and a reduction of cross contamination risks (Leite et al., 2012). Moreover, this is a recognized method to obtaining better RNA yield and quality (Cury and Koo, 2007; Cury et al., 2008; Jahn et al, 2008; Klein et al., 2012; Stipp et al., 2013), due to its capacity of lysing any bacterial cell wall and release all nucleic acids from the cells without degrade them (Leite et al., 2012). Besides, we used a commercial extraction kit based on guanidine thiocyanate salts added of β-ME, which irreversibly denature RNases (Leite et al., 2012), reduces time-consuming and avoid contact with phenol/chloroform (Phongsisay et al., 2007), such as occurring with previous techniques for RNA extraction from oral biofilms (Cury and Koo et al., 2007; Stipp et al., 2008, Falsetta et al., 2012). Gu et al. (2010) and Simón-Soro et al. (2014) were the ones studies found relating RNA extraction from bacteria in human dentine, using lysis buffer associated with sonication or mechanical disruption and commercial kit, however no details about methodology steps, yield and quality of RNA extracted were demonstrated.
Additional RNA purification with RNeasy MinieluteTM CleanUp kit was performed in our research to guarantee the elimination of salt and other possible contaminants (Junttila et al., 2009), as well as to improve RNA yield (Cury and Koo, 2007). The purified RNA samples showed good performance in reverse transcriptase reactions, since the cDNA templates were amplified with efficiency in RT-qPCR assays. These reactions were performed with S. mutans virulence primers (atpD and fabM), responsible for its acidogenic and aciduric properties. Besides, even in low presence in the bacterial population from carious dentine as cited by some authors (Aas et al., 2008; Dige et al., 2014; Lima et al., 2011; Simón -Sóro et al., 2013), S. mutans genes were detected in RT-qPCR, confirming the efficiency of this customized RNA extraction method.
Thus, the cell mechanical disruption associated to commercial kit was considered an acceptable RNA extraction and purification method from carious dentine and, consequently, satisfactory for bacterial gene expression analysis using RT-qPCR. With this finding, bacterial transcriptional researches in human dentinal caries can be well conduced and can promote new knowledge about behavior of bacteria in dentinal sites.
5. Acknowledgments
This study was supported by CNPq (475346/2011-4 -MCT/CNPq 14/2011 Universal).
6. References
Aas, J.A., Griffen, A.L., Dardis, S.R., Lee, A.M., Olsen, I., Dewhirst, F.E., Leys, E.J., Paster, B.J., 2008. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 46, 1407–1417.
Andersson, A.F.,2006. Global analysis of mRNA stability in the archaeon Sulfolobus. Genome Biol. 7, R99.
Benítez-Páez, A., Belda-Ferre, P., Simón-Soro, A., Mira, A., 2014. Microbiota diversity and
gene expression dynamics in human oral biofilms. BMC Genomics. 15, 311.
Bustin, S.A., Benes, V., Nolan, T., Pfaffl, M.W., 2005. Quantitative real-time RTPCR– a perspective. J Mol Endocrinol. 34, 597–601.
Cury, J.A., Koo, H., 2007. Extraction and purification of total RNA from Streptococcus mutans biofilms. Analytical Biochemistry, 365(2), 208–214.
Cury, J.A., Seils, J, Koo, H, 2008. Isolation and purification of total RNA from Streptococcus mutans in suspension cultures and biofilms. Braz Oral Res. 3, 216-22.
Dige, I., Grønkjær, L., Nyvad, B., 2014. Molecular studies of the structural ecology of natural occlusal caries. Caries Res. 48(5), 451-60.
Falsetta, M.L., Klein, M.I., Lemos, J.A., Silva, B.B., Agidi, S., Scott-Anne, K.K., Koo, H.
Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in
Streptococcus mutans to modulate virulence expression in vivo Antimicrob. Agents Chemother. 56(12), 6201-11.
Falsetta, M. L., Klein, M. I., Colonne, P. M., Scott-Anne, K., Gregoire, S., Pai, C. H., Gonzalez-Begne, M., Watson, G., Krysan, D.J., Bowen, W.H., Koo, H., 2014. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infection and Immunity. 82, 1968–1981.
Fejerskov, O., 2004. Changing paradigms in concepts on dental caries: consequences for oral health care. Caries Res. 38(3),182-91.
França, A., Bento, J.C., Cerca, N., 2012a. Optimizing a qPCR gene expression quantification assay for S. epidermidis biofilms: a comparison between commercial kits and a customized protocol. PLoS One. 7(5), e37480.
França, A., Bento, J.C., Cerca, N., 2012b. Variability of RNA quality extracted from biofilms of foodborne pathogens using different kits impacts mRNA quantification by qPCR. Curr Microbiol. 65(1), 54-9.
metatranscriptome of a healthy multispecies biofilm model. J Bacteriol. 194(8), 2082–2095. Geiger, J.R., Melcher, T., Koh, D.S., Sakmann, B., Seeburg, P.H., Jonas, P., Monyer, H., 1995. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and inter-neurons in rat CNS. Neuron. 15,193–204. Gosalbes, M. J., Durban, A., Pignatelli, M., Abellan, J. J., Jimenez-Hernandez, N., Perez-Cobas, A. E., Latorre, A., Moya, A., 2011. Metatranscriptomic approach to analyze the functional human gut microbiota. PLoS ONE. 6, e17447.
Gu, F., Bresciani, E., Barata, T. J., Fagundes, T. C., Navarro, M. F., Dickens, S. H., J.C.
Fenno, Peters, M. C., 2010. In vivo acid etching effect on bacteria within caries-affected
dentin. Caries Research. 44, 472–477.
Holt, J.G., Krieg N.R., Sneath P.H.A., Staley J.T., Williams S.T.,1994. Bergey’s Manual of Determinative Bacteriology. Williams & Wilkins. 9 Edition. Baltimore.
Imbeaud, S., Graudens, E., Boulanger, V., Barlet, X., Zaborski, P., Eveno, E., Mueller, O., Schroeder A., Auffray, C., 2005. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Research. 33, 1–12.
Jahn, C.E., Charkowski, A.O., Willis, D.K.J, 2008. Evaluation of isolation methods and RNA integrity for bacterial RNA quantitation.Microbiol Methods. 75(2), 318-24.
Jeon, J.G., Pandit, S., Xiao, J., Gregoire, S., Falsetta, M.L., Klein, M.I., Koo, H., 2011. Influences of trans-trans farnesol, a membrane-targeting sesquiterpenoid, on Streptococcus mutans physiology and survival within mixed-species oral biofilms. Int J Oral Sci. 3, 98-106. Junttila, S., Lim, K.J., Rudd, S., 2009. Optimization and comparison of different methods for RNA isolation for cDNA library construction from the reindeer lichen Cladonia rangiferina. BMC Res Notes. 2, 204.
Kempf, P.F., Lee, S., Laroche, J., 1993. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl Environ Microbiol. 59, 2594–2601.
Klein, M.I., DeBaz, L., Agidi, S., Lee, H., Xie, G., Lin, A. H.M., Hamaker, B.R., Lemo, J.A., Koo, H., 2010. Dynamics of Streptococcus mutans transcriptome in response to starch and sucrose during biofilm development. PLoSONE. 5, e13478.
Klein, M.I., Scott-Anne, K.M., Gregoire, S., Rosalen, P.L., Koo, H., 2012. Molecular approaches for viable bacterial population and transcriptional analyses in a rodent model of dental caries. Mol. Oral Microbiol. 27, 350– 361.
Leite, G. M., Magan, N., Medina, Á., 2012. Comparison of different bead-beating RNA extraction strategies: An optimized method for filamentous fungi. Journal of Microbiological Methods. 88, 413–418.
Ludwig, W., Schleifer, K.H., 2000. How quantitative is quantitative PCR with respect to cell counts? Syst Appl Microbiol. 23(4), 556-62.
Nyvad, B., Crielaard, W., Mira, A., Takahashi N., Beighton, D., 2012. Dental caries from a molecular microbiological perspective. Caries Res. 7(2), 89-102.
Nolan, T., Hands, R.E., Bustin, S.A., 2006. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 1: 1559–1582.
Nour, A.M., Barbour, E.K., Depint, F., Dooms, M., Niang, K., 2010. Comparison of five RNA extraction methods from rabbit’s blood. Agriculture and Biology Journal of North America. 1: 448–450.
Peterson, S.N., Meissner, T., Su, A.I., Snesrud, E., Ong, A.C., Schork, N.J., Bretz, W.A., 2014. Functional expression of dental plaque microbiota. Front Cell Infect Microbiol. 14;4:108.
Raeymaekers, L., 1993. Quantitative PCR — theoretical considerations with practical implications. Anal. Biochem. 214, 582–585.
Rump, L.V., Asamoah, B., Gonzalez-Escalona, N., 2010. Comparison of commercial RNA extraction kits for preparation of DNA-free total RNA from Salmonella cells. BMC Res Notes. 3, 211.
Selinger, D.W., Saxena, R.M., Cheung, K.J., Church, G.M., Rosenow, C., 2003. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 13, 216 –223.
Simón-Soro, A., Belda-Ferre, P., Cabrera-Rubio, R., Alcaraz, L.D., Mira, A., 2013. A tissue-dependent hypothesis of dental caries. Caries Res. 47(6), 591-600.
Simón-Soro, A., Guillen-Navarro, M., Mira A., 2014. Metatranscriptomics reveals overall active bacterial composition in caries lesions. Journal of Oral Microbiology. 6, 25443.
Stipp, R.N., Gonçalves, R.B., Höfling, J.F., Smith, D.J., Mattos-Graner, R.O., 2008. Transcriptional analysis of gtfB, gtfC, and gbpB and their putative response regulators in several isolates of Streptococcus mutans. Oral Microbiol Immunol. 6, 466-73.
Stipp, R.N., Boisvert, H., Smith, D.J., Höfling, J.F., Duncan, M.J., Mattos-Graner, R.O., 2013. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS One. 8(3), e58271.
Twin, J., Bradshaw, C. S., Garland, S. M., Fairley, C. K., Fethers, K., Tabrizi, S. N., 2013. The potential of metatranscriptomics for identifying screening targets for bacterial vaginosis. PLoS ONE. 8(9), e76892.
Table 01. RNAyield, purity and gene expression values of S. mutans from dentine lesions (n=14)
Yield (ng/µl) Ratio (A260/A280) atpD gene expressiona fabM gene expressionb
67.01 ± 22.33 2.00 ± 0.07 0.007 ± 0.010 0.007 ± 0.011
Values are means ± standard deviation. cDNA mass used to begin the RT-qPCR (20 ng) was used for normalization of gene expression.
Fig. 2. RT-qPCR amplification plot and standard curve data for atpD gene from S. mutans. The values of expression detected for the tested samples (depicted in blue) are within the standard curve range.
Slope= -3.35; Y-Intercept= 19.48; Correlation coefficient (R2)= 0.998; Efficiency %= 98.862
CAPÍTULO 2
Virulence gene expression of Streptococcus mutans in active dentine lesions from
children with early childhood caries
D.S. Bezerraa, R.N. Stippb, B.G. Nevesa, S.F.F. Guedesa, L.K.A. Rodriguesa
a
Post-graduation Program, Faculty of Pharmacy, Dentistry and Nursing, Federal University of Ceará, Fortaleza, Ceará, Brazil; bDepartment of Microbiology and Immunology, Piracicaba Dental School, State University of Campinas, Piracicaba, SP, Brazil
Short Tittle: S. mutans virulence gene expression in dentine caries.
Key Words:Dentine Caries; Early Childhood Caries; Streptococcus mutans; RNA Extraction; Gene Expression
Address all correspondence to:
Lidiany K. A. Rodrigues, DDS, MSc, PhD, Associate professor
Postgraduate Program in Dentistry, Faculty of Pharmacy, Dentistry and Nursing Federal University of Ceará, Fortaleza, Ceará, Brazil.
Rua Monsenhor Furtado - Zip Code: 60430-355 Phone- #+558533668410/ Fax- #+558533668232 Email: lidianykarla@yahoo.com
Conflict of interests
There are no potential conflicts of interest for any of the authors.
ABSTRACT
Streptococcus mutans (SM) are organisms strongly associated with the development of dental caries, thus their genetic and phenotypic responses were investigated in active dentinal lesions. This study aimed to verify the prevalence and quantity of viable SM as well as to analyze their expression of virulence genes in active and arrested dentine lesions of children with early childhood caries (ECC). Dentine samples from 29 active and 16 arrested lesions were harvested from pre-school children (age 2-5 years). Total RNA was extracted and the RT-qPCR reactions were performed for SM identification, quantification and analysis of virulence gene expression (atpD, fabM, nox, pdhA and aguD). Although expressed in low quantity in relation to total streptococci (TS) and total bacteria (TB), no statistically significant differences were found for SM prevalence and abundance in active and arrested caries lesions (p>0.05). SM expressed all studied virulence genes in both groups, but pdhA
Introduction
With prevalence as high as 70% in developing countries as well as underprivileged populations in developed ones [King and Wong, 2006], dental caries can contribute to bad impact in quality of life of children including pain, weight loss and a poor scholar performance [Filstrup et al., 2003; Feitosa et al., 2005; Abanto et al., 2011]. Some bacteria, such as Streptococcus mutans (SM), play key roles as agents responsible for the onset and progression of dental caries, including early childhood caries (ECC) [Mattos-Graner et al., 1998, 2001; Nobre-dos-Santos et al., 2002; Aas et al., 2008; Takahashi and Nyvad, 2008]. SM virulence can be attributed to the production of organic acids from sugar metabolism (acidogenicity), enhanced tolerance to environmental stress, particularly low pH (acidurance) [Hazlet et al., 1999; Quivey et al., 2001; Lemos et al., 2005; Sztajer et al., 2008] and capacity of forming biofilm on smooth tooth surfaces, which are important properties for enamel demineralization, one initial stage of caries lesions [Bowen and Koo, 2011].
Several molecular based diversity studies have reported the presence of mutans streptococci in dentine caries [Aas et al., 2008; Lima et al., 2011; Belda-Ferre et al., 2012; Kuribayashi et al., 2012; Wolff et al., 2012; Simón-Soro et al., 2013; Obata et al, 2014], including as being part of the metabolically active bacterial composition of dentine lesions [Simón-Soro et al., 2014]. However, despite of being frequently associated to caries, the impact of SM persistence in caries dentine activity is not clear. Therefore, it is important not only to describe the bacteria involved in caries processes, but also to know how bacteria are acting [Vieira et al., 2012], since immunological, environmental and genetic factors can influence caries lesion progression. Thus, an understanding of how SM deals with different environments, analyzing site-specific samples is essential to comprehend its cariogenicity being important for designing examples of interventions to eliminate or reduce the proportions of cariogenic organisms in advanced lesions.
Several genes identified and studied over the years can modulate acidogenicity and acidurance of bacteria [Kuramitsu et al, 1993; Banas et al., 2004]. For SM, the membrane-bound F1F0-ATPase system, which pumps protons from cells while maintaining the internal pH value, is the primary determinant of acid production and tolerance, and are encoded by
NAD+ and the maintenance of NAD+/NADH ratio (nox gene) [Derr al., 2011] and production of alkalis by the agmatine deiminase system (which has aguD gene) [Burne and Marquis, 2000], which reinforce the competitive fitness of SM, contributing to its persistence and pathogenesis [Griswold et al., 2004, 2006]. Furthermore, in situations of starved glucose, aiming to maintain the viability of bacterial cells, some genes for metabolism regulation are expressed, as pdhA gene bound to the regulation of pyruvate dehydrogenase pathway [Busuoic et al., 2010; Moye et al., 2014].
Active dentinal lesions exhibit evidence of progression or change over time, being more aggressive and presenting more possibilities of causing serious damage to children quality of life [Filstrup et al., 2003]. On the other hand, non-progressive arrested lesions, featured by changes in color (darkness) and consistency in surface layer (leather to hard) [Nyvad et al. 1997], exhibit higher micro-hardness and slow progression [Chu and Lo, 2008]. In this way, bacterial viability and virulence gene expression could be influenced by modifications of intrinsic environmental characteristics [Quivey et al., 2001; Lemos et al., 2005; Wen et al., 2010; Moye et al., 2014].
Thus, the objective of this research was to investigate, for the first time, the prevalence and quantification of viable S. mutans (SM) in relation to total streptococci (TS) and total bacteria (TB) in active and arrested cavitated dentin lesions of children with ECC, as well as its expression of virulence genes linked with acidogenicity and acidurance.
Material and Method
Study population
the end, 40 children participated in this research, because 14 were not able to attend two or more scheduled appointments, 08 had insufficient quantity dentine due very small exposed surface, 05 were sick in the period of sample collection and 02 had an aggressive behavior at the moment of collection. The study design, protocol, and informed consent were approved by the Ethics Committee of the Federal University of Ceará, protocol number 548.405.
Clinical examination
Dentine samples were collected in a clinic at the Dental School of Federal University of Ceará, Brazil. In the first visit, a clinical examination was done by calibrated researchers using the visual criteria International Caries Detection and Assessment System II (ICDAS II) [Pitts et al., 2004; Ismail et al., 2005] associated to the Nyvad System for differentiation between active and arrested caries [Nyvad et al., 1999]. The samples selected for our study were included in score 5 (distinct cavity with visible dentine) or 6 (extensive - more than half the surface - distinct cavity with visible dentine) from ICDAS II. For Nyvad System, the scores 3 (active dentine caries with whitish/yellowish surface, feels soft or leathery on gentle probing) or 6 (inactive dentine caries with brown/ black and shiny surface and feels hard on probing with gentle pressure) were adopted [Nyvad et al., 1999]. Radiographic exams and treatment plans were made and the follow-up office visit was scheduled.
Sample Collection
samples were stored in to sterile and RNAse-free microtubes (Axygen, Union City, CA, USA) containing RNA stabilizer solution (RNAlaterTM - Ambion Inc., Austin, TX, USA) and stored at 4°C for 18 h. After this period, the tubes were transferred to freezer (-80oC) for storage until RNA extraction.
Cavities were lined and restored with a suitable material. At the end of the procedures, the children and their parent or guardian were instructed about oral hygiene and healthy eating habits. Patients were scheduled for other appointments if these were needed.
RNA extraction and purification
The dentine samples were thawed and centrifuged (11,000 × g/ 1 min/ 4°C). The
RNAlater TM solution was removed using an automatic pipettor without disturbing the pellet. The samples were carefully transferred to cryogenic tubes containing 0.16 g of 0.1 mm diameter zirconium beads (Stipp et al., 2013). The mechanical disruption of bacterial cells was made by Mini-Beadbeater (Biospec Products Inc., Bartlesville, OK, USA) at maximum power (2 cycles of 60 s with 1 min rest on ice). Then, 850 �l of RLT buffer (Qiagen, Valencia, CA, USA) with 10% of β-mercaptoethanol (β-ME) were added and the suspension homogenized by vortexing. After centrifugation (11,000 × g/ 2 min/ 4°C), aliquots of 350 �l
of the supernatant were subjected to extraction with RNeasy Mini KitTM according the manufacture’s instructions. The RNA concentration and purity (absorbance ratio A260/A280 and
A260/A230) were measured using Nanodrop 2000c spectrophotometer (Thermo Scientific, Wilmington, DE). The RNA integrity was verified by visualization of 16S and 23S ribosomal RNA bands on 1.2% formaldehyde-agarose gel stained with ethidium bromide. Next, the samples were treated with TurboTM DNAse (Applied Biosystems, Ambiom, Austin, TX, USA) for removal of genomic DNA. RNA solution was again purified using RNeasy MinieluteTM CleanUp kit (QUIAGEN, Dus, Bundesland, Germany) performed according to the manufacturer’s recommendations. The RNA concentration, purity and integrity were obtained again. The later samples were stored at -80°C.
cDNA synthesis
free-water to complete 30 µl of solution, which were incubated at 25°C for 5 min, 42°C for 2 hours and finally 85°C for 5 min. Next, 70 µl of nuclease free water were added to each tube to obtain a normalized solution of 10 ng/µl of cDNA.
Sanger sequencing
Six cDNA samples were submitted to conventional PCR with primers for SM virulence genes (table 2) and subsequently to Sanger sequencing. The objective was to investigate specificity of the primers for SM genes in human samples, a purpose they were not previously credited. In addition, 16S rRNA gene for SM (5’- ACCAGAAAGGGACGGCTAAC – 3’and 5’- TAGCCTTTTACTCCAGACTTTCCTG – 3’) [Klein et al., 2010] was also sequenced because its expression could be used for normalizing the levels of genes’ expressions. Primers for bacteria identification (table 1) did not had their specificity investigated because they were validated for use in human samples previously [Nadkarni et al., 2002; Yano et al., 2002; Sakagushi et al., 2010]. PCR assays were executed in a Veriti® Thermal Cycler (Applied Biosystems, Foster City, CA, USA) and the reaction mix included 1 µl of Buffer PCR 10x, 1 µl of dNTPs 10mM, 2.5 µl of MgCl2, 3 µl of each primer F/R 10 µM, 0.25 µl of Taq polymerase 5 U/µl, 50 mM 1 µl of template cDNA (10 ng/µl) and 37.25 µl of nuclease-free water. The thermal-cycling consisted in 5 min at 95°C; 35 cycles of 15 s at 95°C, 60 s at melt temperature of each primer (table 2), 20 s at 72°C; 4 min at 60°C). Their products were purified with commercial kit (QIAquick® PCR Purification Kit, QUIAGEN, USA) and the sequencing reactions were performed using Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, USA) and submitted to thermal-cycling (20 s at 95°C; 40 cycles of 15 s at 96°C, 15 s at 50° C and 4 min at 60°C; 60s at 60°C). At the end, the products were precipitated, dried, resuspended in 10 µl of formamide and denatured for 5 min at 95°C. Sanger sequencing occurred in a 3500 Series Genetic Analyzer (8-capillary) (Applied Biosystems, Foster City, CA, USA). The sequencing data obtained was submitted to BioEdit software, version 7.2.5, to obtain contigs sequences, which were submitted to analysis using BLAST program (Basic Local Alignment Search Tool), available at http://estexplorer.biolinfo.org/hsd, to verify sequences similarity.
In order to get DNA from S. mutans UA159 for obtain standards curves, this strain was cultured in broth for 24 h as recommended by Bergey’s Manual of Determinative Bacteriology (Holt et al. 1994). After centrifugation and washing in sterile saline solution (sodium chloride 0.9%), the quality and purity of bacterial cultures were checked by Gram staining and their DNA was recovered using an organic extraction protocol based on phenol/chloroform purification and alcohol precipitation (Wilson, 2001). Serial dilutions starting from 600 ng to 0.0003 ng (10-fold) of S. mutans DNA concentrations were used as standards and positive controls for relative quantification of the targeted bacteria. A standard amplification curve and a melting-point product curve were obtained for each primer set.
The RT-qPCR assays were performed to test the functionality of RNA extracted in gene expression studies using a StepOneTM Real Time PCR System (Applied Biosystems, Foster City, CA, USA). The reactions were performed using MicroAmpFast Optical 48-Well Reaction Plate (Applied Biosystems, Foster City, CA, USA) covered with Optical Adhesive Film (Applied Biosystems). A mixture of 5 µl of Power SYBR® Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA), 2.4 µl of nuclease-free water, 0.3 µl of each primer F/R 10 µM and 2 µl of cDNA (20 ng) was added in each spot of a 48-well plate. Assays were carried out in duplicate, and the final analyses were based on the mean of the two reactions. Negative control included reactions without template. The standard curves were used to transform the quantification cycle (Cq) values to the mass of cDNA amplified. The normalization of gene expression was made using the cDNA amount initially inserted in each spot of 48 well-plate (20 ng).
Statistical Analyses
Inc., Chicago, IL, USA) was used to perform the analyses. The level of significance was set at 5%.
Results
All sequenced PCR products were 100% compatible only for S. mutans (SM) according Sanger sequencing, except 16S rRNA gene, which showed similarity to other sequences in the database such as Streptococcus sanguinis; Streptococcus gordonii; Streptococcus oralis and Streptococcus sobrinus.
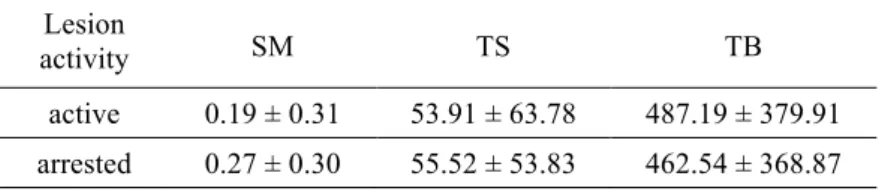
Viable SM and total oral streptococci (TS) were identified in dentinal samples and no statistically significant differences were found for bacterial prevalence in active and arrested caries (table 3). Table 4 shows absolute quantification of metabolically active bacteria in active and arrested carious samples. There were no statistically significant differences for SM, TS and TB (p>0.05). The quantification of S. mutans in relation to total streptococci (SM/TS) and in relation to total bacteria load (SM/TB), as well as the abundance of TS in relation to TB (TS/TB) was illustrated in figure 2. TS/TB were more abundant in the arrested dentin lesions (p=0.025) than in active, while SM/TS and SM/TB did not show correlation with the lesion status.
Figure 3 shows the SM virulence gene expression in active and arrested dentine caries lesions. Only pdhA (p=0.04)and aguD (p=0.05) genes were more expressed in arrested caries. The others did not present significant statistically differences (p>0.05).
Discussion
[2013], while this may occurs due to the deficiency of current molecular methods in dealing with very small bacterial samples, such studies probably will not increase knowledge and detailed insight about the microbial and metabolic site-specific processes of caries.
For detecting and quantifying metabolically active microbial members and their genes expression, mRNA analysis is an ideal alternative [Nyvad at al., 2013]. In fact, it has been postulated that this analysis can be more relevant for explaining caries activity than studies focusing exclusively on the microbiome [Takahashi and Nyvad, 2008], however these studies are not frequent in the literature. The limitation of this approach comes from the high percentage of rRNA present in bacterial samples, which typically accounts for over 90% of total RNA [Gosalbes et al., 2011]. In our study, to increase the chances of success, we used a standard and previously tested technique of RNA extraction to overcome these difficulties.
For TB, our quantification results differed from previous microbiological studies, which observed a higher concentration of bacteria in soft/leathery (active) caries lesions than in hard/dark (arrested) lesions [Schupback et al. 1995; Schete et al., 1996]. However, these studies focused in bacterial culture media, method less reliable in relation to molecular methods. On the other hand, our results showed a higher relative abundance of TS load in arrested lesions, reaching 16.95% of total bacteria, which was previously found in dentine lesions, regardless of caries activity, from children [Jiang et al., 2014] and adults [Simón-Soro et al., 2013]. It can be suggested a possible contribution of these microorganisms to the inactivation of the lesions, since several streptococcus are able to produce alkalis by the arginine deiminase system (ADS) [Nascimento and Burne, 2014]. These bacteria generates ammonia that neutralizes acids in pathogenic biofilms and favors pH increase, which are compatible with the pH of arrested lesions as previously demonstrated [Hojo et al., 1994; Kuribayashi et al., 2012]. Other point to be considered is that arrested lesions present a harder reactive dentine with irregularly and blocked dentinal tubules [Love and Jenkinson, 2002] and lower water and protein contents [Nanci, 2012]. In this way, proteolytic bacteria, generally higher in active dentinal caries [Simón-Soro et al., 2013], seem do not have adequate nutritional conditions for their survival causing the proliferation of more acidogenic organisms such as species of streptococcus.