ContentslistsavailableatScienceDirect
International
Journal
of
Biological
Macromolecules
jo u r n al h om ep age :w w w . e l s e v i e r . c o m / l o c a t e / i j b i o m a c
Structural
characterization
of
two
isolectins
from
the
marine
red
alga
Solieria
filiformis
(Kützing)
P.W.
Gabrielson
and
their
anticancer
effect
on
MCF-7
breast
cancer
cells
Renata
Pinheiro
Chaves
a,
Suzete
Roberta
da
Silva
a,
Luiz
Gonzaga
Nascimento
Neto
a,b,
Romulo
Farias
Carneiro
a,
André
Luis
Coelho
da
Silva
c,
Alexandre
Holanda
Sampaio
a,
Bruno
Lopes
de
Sousa
d,
Maria
Guadalupe
Cabral
e,
Paula
Alexandra
Videira
f,
Edson
Holanda
Teixeira
b,
Celso
Shiniti
Nagano
a,∗aLaboratóriodeBiotecnologiaMarinha–BioMar-Lab,DepartamentodeEngenhariadePesca,UniversidadeFederaldoCeará,CampusdoPicis/n,bloco
871,60440-900Fortaleza,Ceará,Brazil
bLaboratórioIntegradodeBiomoléculas–LIBS,DepartamentodePatologiaeMedicinaLegal,UniversidadeFederaldoCeará,MonsenhorFurtado,s/n,
60430-160Fortaleza,Ceará,Brazil
cLaboratóriodeBiotecnologiaMolecular–LabBMol,DepartamentodeBioquímicaeBiologiaMolecular,UniversidadeFederaldoCeará,CampusdoPici,
bloco907,60440-900,Fortaleza,Ceará,Brazil
dFaculdadedeFilosofiaDomAurelianoMatos,UniversidadeEstadualdoCeará,Av.DomAurelianoMatos,2060,LimoeirodoNorte,CE,62930-000,Brazil eCEDOC,NOVAMedicalSchool,UniversidadeNOVAdeLisboa,1150-082,Lisbon,Portugal
fUCIBIO,DepartamentodeCiênciasdaVida,FaculdadedeCiênciaseTecnologia,UniversidadeNOVAdeLisboa,2829-516,Caparica,Portugal
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received21July2017 Accepted28September2017 Availableonline29September2017
Keywords: Marinealga Lectin Anticancereffect
a
b
s
t
r
a
c
t
Asdescribedintheliterature,Solieriafiliformislectin(SfL)fromthemarineredalgaS.filiformiswasfound tohaveantinociceptiveandanti-inflammatoryeffects.Inthisstudy,wecharacterizedtwoSfLvariants, SfL-1andSfL-2,withmolecularmassof27,552Daand27,985Da,respectively.Theprimarystructures ofSfL-1andSfL-2consistoffourtandem-repeatproteindomainswith67aminoacidseach.SfL-1and -2showedhighsimilaritytoOAAH-familylectins.3DstructurepredictionrevealedthatSfL-1and-2are composedoftwo-barrel-likedomainsformedbyfiveantiparallel-strands,whichareconnectedbya shortpeptidelinker.Furthermore,themixtureofisoforms(SfLs)showedanticancereffectagainstMCF-7 cells.Specifically,SfLsinhibited50%ofviabilityinMCF-7cellsaftertreatmentat125g.mL−1,while theinhibitionofHumanDermalFibroblasts(HDF)was34%withthesametreatment.Finally,24hafter treatment,25%ofMCF-7cellswereinearlyapoptosisand35%inlateapoptosis.Evaluationofpro-and anti-apoptoticgeneexpressionofMCF-7cellsrevealedthatSfLsinducedcaspase-dependentapoptosis within24h.
©2017ElsevierB.V.Allrightsreserved.
1. Introduction
Lectins,whichareubiquitouscarbohydrate-bindingproteinsor glycoproteins,havereceivedspecialattentionincancerprognosis, diagnosis,andtreatmentbytheirbiologicalactivities.Lectinscanbe usedastoolstoidentifyaberrantglycansonthemembranesurface ofneoplasiccellsandasantitumormoleculestoinduceapoptosis andautophagythroughvariousmechanisms[1,2].
∗ Correspondingauthorat:BioMar-Lab,DepartamentodeEngenhariadePesca, UniversidadeFederaldoCeara,Av.HumbertoMontes/n,60440-900,Fortaleza, Ceara,Brazil.
E-mailaddress:naganocs@gmail.com(C.S.Nagano).
Marine algaeare good sources of newlectins. Algae lectins possess unique molecular structures and important biological activities.Inparticular,theirhighspecificityforcomplex carbohy-dratesandglycoconjugatesmakesthemusefulforbiochemicaland biologicalapplications[3,4].Biochemicalstudiesrevealthatmany lectinsisolatedfrommarineredalgaesharecommonproperties withprokaryotelectins[5].Theselectinscouldbegroupedintoa familyoflectinsknownasOscillatoriaagardhiiagglutinin homo-logue(OAAH)lectinfamily,amongwhichisalectinisolatedfrom thecyanobacteriumO.agardhii[6].
OAA, which contains 132 residues with two tandem-repeat domains[7],issimilartoPFAfromthebacteriumPseudomonas fluo-rescens[8].Ontheotherhand,BOAfromthebacteriumBurkholderia oklahomensis EO147[9],MBHA fromthebacteriumMyxococcus
Xanthus[10],ESA-2fromthemarinealgaEucheumaserra[11], EDA-2fromthemarinealgaEucheumadenticulatum[12],KSA-2from themarinealgaKappaphycusstriatum[13],andKAA-1/-2fromthe marinealgaKappaphycusalvarezii[14]allcontainfour tandem-repeatsequences246–269residuesinlength.
OAAH-family members share high identity of amino acid sequenceandpossessduplicateorquadruplicatedomainsintheir sequencesthatleadtosingular-barrel-liketopologyinvolvedin thetightbindingtooligomannosides[7,15].Interestingly,amino acidsequencesofOAAHlectinsarecompletelydistinctfromother highmannose-bindinglectins.
SolieriafiliformisisamarineredalgafoundalongtheBrazilian northeastcoast.Itslectin,SfL,at29kDawasisolatedbyBenevides andcollaborators[16].Abreuandcoauthors[17]showedthatSfL enhancestheproductionofIL-6athighlevels,butwithno cyto-toxicactivityonsplenocytes,thuspromotinganti-inflammatory effect.Inaddition,SfLseemstohaveimportantinvivo antinocicep-tiveactivityandcouldrepresentapotentialtherapeuticagentfor invitroandinvivoimmunomodulatoryeffects[18].However,other biologicalactivitieshavenotbeenexploited,includingthislectin’s antitumoractivity.
Breastcancer,theforemostcauseofdeathinwomen,isa het-erogeneousdiseasecharacterizedbya varietyofmolecularand geneticalterationsthatinducegrowthandsurvival[19,20]. Cur-rently,chemotherapyisthemostcommonlyusedmethodtotreat cancer.However,chemotherapeutictechniquesarelimitedbytheir highcellulartoxicity,aswellastheoccurrenceofsideeffects[21]. Therefore,selectivebiologicaltreatmentshaveemergedtotreat certaintypesofcancersortotargetspecificdeterminantsexpressed bymanydifferenttumors[22].
Inthiswork,weidentifiedandthenconductedstructuralstudies ontwoisolectinsofthemarineredalgaS.filiformis:SfL-1and SfL-2.Theirprimarystructuresweredetermined,theirtridimensional structureswerepredicted,andmoleculardockingwasperformed. ThecytotoxiceffectoftheisolectinsonMCF-7cellswasalso eval-uated.
2. Methods
2.1. Algaecollection
SpecimensfromthemarineredalgaSolieriafiliformis(Kützing) P.W.GabrielswerecollectedintheintertidalzoneofPachecoBeach, Ceará,Brazil.Algaeweretransportedtothelaboratoryiniceand keptat−20◦C untiluse. Asmall portionof thespecimens was storedat−80◦CforRNAextraction.Collectionswereauthorized
andcertifiedbyresponsibleenvironmentalinstitutions(SISBIOID: 33913-8).
2.2. Purificationoflectin
ThelectinfromthemarineredalgaS.filiformis(SfL)waspurified followingtheprotocoldescribedbyBenevidesandcollaborators [16]withminormodifications.Frozenalgaeweregroundtoafine powderbymortarandpestleinthepresenceofliquidnitrogen. Thepowderwasstirred(1:3w/v)with20mMphosphatebuffer, pH7.0,containing150mMNaCl(PBS),for4hatroomtemperature. Insolublealgaematerialwasremovedbycentrifugationat5000×g for30minat4◦C.Thesupernatant(crudeextract)wascollectedand assayedforhemagglutinatingactivityand proteinconcentration [23].
Thecrude extract wassubmitted toprecipitation withsolid ammoniumsulfate(70%ofsaturation)for4handcentrifugedat 5000×gfor30minat4◦C.Theresultingprecipitate(F0/70)was recoveredinPBSanddialyzedagainstdeionizedwaterandthen
against20mMphosphatebuffer,pH7.0(PB).Aftercentrifugation, F0/70wasappliedtoaDEAE-SephacelColumn(1.0cm×3.0cm)
previouslyequilibratedwithPB.Thecolumnwaswashedwiththe samebufferataflowrateof1mLmin−1untilthatcolumn’s
efflu-entshowedabsorbanceat280nm<0.02(P1).Theadsorbedfraction
(P2)waseluted withPBcontaining1MNaCl.Two-mLfractions
werecollectedandtestedforhemagglutinatingactivityagainst3% trypsin-treatedrabbiterythrocytes.Fractionsshowing hemagglu-tinatingactivity(P1)werepooled,dialyzedagainstdistilledwater,
andfreeze-dried.
2.3. Molecularmassdetermination
The isotopicaveragemolecularmass of SfLwasdetermined byliquidchromatographycoupledtoElectrosprayionization-mass spectrometry(LC-ESI–MS)usingahybridSynaptHDMSmass spec-trometer (Waters Corporation, MA, USA). Two g of SfL were appliedtoaC-18nanocolumn(75m×100mm)andelutedwith
gradientof10% to85% ofACNcontainingformicacid(FA)0.1% ataflowrateof0.6Lmin−1.Eluatesweredirectlyinfusedinto themassspectrometerusingananoAcquitysystemconnectedtoa nanoelectrospraysource.
The instrument was calibrated with [Glu1]-fibrinopeptide-B collision-induceddissociation(CID)fragments.Massspectrawere acquiredbyscanningatm/zrangingfrom800to3000at1scan s−1.Themassspectrometerwasoperatedinpositivemode,using asourcetemperatureof363Kandcapillaryvoltageat3.5kV.Data collectionandprocessingwerecontrolledbyMassLynx4.1 soft-ware (WatersCorporation,MA, USA).The deconvolution of ESI massspectrawasperformedusingtheMaxEnt1algorithminthe MassLynxsoftware.
2.4. Primarystructuredetermination
2.4.1. N-terminalaminoacidsequencing
The N-terminal aminoacidsequence of SfLwasdetermined by Edmandegradation ina Shimadzu model PPSQ-31Aprotein andpeptidesequencer(ShimadzuCorp.,Japan).PTH-aminoacids fromtheN-terminussequencewereseparatedona2.0×250mm
WakosilODS column(WakoPureChemicalCorp.,Osaka,Japan) connectedtoamodelLC-20ATpump.Theabsorbancewasdetected at269nmwithaUV–visSPD-20Adetector.
The sequence similarity of the N-terminus was evaluated online (http://www.ncbi.nlm.nih.gov/BLAST/), and homologous sequenceswereidentifiedwiththebasiclocalalignmentsearch toolprogram(BLAST)usingPROTEINBLASTfromtheNational Cen-terforBiotechnologyInformation(NCBI).
2.4.2. Tandemmassspectrometry(MS/MS)ofpeptides
SDSPAGEwasperformedasdescribedbyLaemmli[24].After staining, SfL spots were excised as described by Shevchenko and collaborators [25]. Discolored spots were subjected to digestion withtrypsin (Promega, Madison, WI, USA). Digestion was performed in ammonium bicarbonate 25mM at 1:50w/w (enzyme/substrate).Thedigestionwasmaintainedat37◦Cfor16h and then was stopped with2L of FA2%. The peptides were extracted,separatedbyreversephasechromatographyinaC-18 column,asdescribed above,and directlyinjectedintothemass spectrometer.
evaluatedonlinebyPROTEINBLASTfromNCBItoidentify homol-ogoussequences.
2.4.3. RapidamplificationofcDNAends(RACE)
TotalcellularRNAfromS.filiformiswasisolatedwithAxyPrep MultisourceTotalRNAMiniprepkit(AxygenBiosciences,CA,USA), accordingtothemanufacturer’sprotocol.First-strandcDNAwas synthesized from total RNA using MMLV RT (Moloney Murine LeukaemiaVirusReverseTranscriptase;Sigma-Aldrich,MO,USA) andadaptorQt(5′-CCAGTGAGCAGAGTGACGAGGACTCGAGCT
CAAGCT(T)16-3′(Sigma-Genosys,TX,USA),accordingtothe
man-ufacturer’sinstructions.
cDNAwasPCR-amplifiedusingone-tenthoftheRTreactionas atemplate.Adegenerateprimerwasdesigned(primer-SfL3′;5′ -GTICAGAATCARTGGGGIGG-3′)basedontheaminoacidsequences obtainedbyMS/MS:VQNQWGG.PCRforamplificationof3′RACE (rapidamplificationcDNA3′ ends)wascarriedoutbyusingTaq DNApolymerase(ThermoScientific,MA,USA).Thereaction con-sistedofatotalvolumeof25L,containing2LofcDNA,0.4M ofeachprimer,0.4mMofdNTPmix(ThermoScientific,MA,USA), and1Uofenzymein1XPCRBufferwith3mMofMgCl2.PCRwas
performedwithprimer-SfL3′andQi(5′ -GAGGACTCGAGCTCAAGC-3′).Amplificationprotocolincludedaninitialdenaturationstepat 95◦Cfor5min,followedby35cyclesofdenaturationat95◦Cfor 30s,annealingat50◦Cfor30s,extensionat72◦Cfor1min,and thefinalextensionstepat72◦Cfor5min.Then,thereactionwas quenchedto4◦Candanalyzedbyagarosegelelectrophoresis.
ThePCRproductwaspurifiedbyillustraTMGFXPCRDNAand
GelBand Purification kit (GE Healthcare, IL,USA). Purified PCR productwascloned intopGEM-T-easy vector, transformedinto Escherichiacoli strainDH5␣(Novagen,Germany), and screened with blue-white selection in LB agar, containing 100gmL−1
ampicillin (Thermo Scientific, MA, USA), 0.5mM IPTG (Iso-propyl- D D-1-thiogalactopyranoside; Thermo Scientific, MA, USA), and 80gmL−1 X-Gal (5-bromo-4-chloro-3-indolyl--d
-galactopyranoside;ThermoScientific,MA,USA).Thecloningwas performedinabiosafetylaboratorycertifiedinaccordancewith governmentalrequirements(CQB:R007-2016).Therecombinant plasmidswereextractedbyillustraTMplasmidPrepMiniSpinkit
(GEHealthcare,IL,USA)and confirmedbydigestionwithEcoRI (Promega,WI,USA).
Finally, constructions were sequenced in an automatic sequencer(MegaBACE;GEHealthcare,IL,USA),usingtheprimers T7promoter(5′-TAATACGACTCACTATAGGG-3′)andSP6(5′-ATT TAGGTGACACTATAG-3′).Sequencingwasperformedwithatleast tenclonesfromPCRamplification.Thereadswereanalyzedbythe Phred-Phrap-Consedprogram[26–28].Thecontigsformedwere translatedwiththeExpasytranslationtool(http://web.expasy.org/ translate/).Aminoacidsequencealignmentswereperformedusing MultAlin[29]andESPript2.2[30].ThesequencesimilarityofSfL wasevaluatedonlinebyNCBI’sPROTEINBLAST.
2.5. StructuralanalysisofSfLisoforms
2.5.1. Proteinmodeling
StructuralpredictionofisoformsIandIIfromS.filiformislectin (SfL-1andSfL-2)wasperformedintheMODELLERprogram,suite v.9.16,usingthestructureofB.oklahomensisagglutininasthe tem-plate(BOA,PDBID4GK9)[9,31].Forstructuralmodeling ofSfL isoforms,MODELLERdefaultparameterswereused,andsequence alignmentcorrectionsweremanuallyedited.Initially,20 theoreti-calmodelsforeachisoformweregenerated,whichwereranked basedontheirDOPE(discreteoptimizedproteinenergy)scores [32].
The best modelswere then selected and analyzed for their stereochemicalproperties(Ramachandranplots, stericoverlaps,
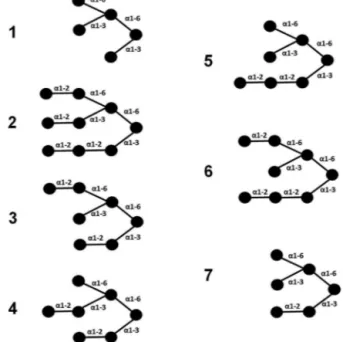
Fig.1. Differentbranchedoligosaccharides.Oligosaccharidesselectedonthebasis ofpreviouslyreporteddataforhomologouslectins.Thecirclesrepresentmannosyl residues.
Cdeviationparameters,rotamers,andbondanglequality)with theMolProbityServer[33].Afterwards,thefinalstructureswere manuallyinspectedusingPyMolMolecularGraphicsSystem, Ver-sion1.8(Schrödinger,LLC).
2.5.2. Moleculardockingcalculations
BindingofSfLisoformstodifferentbranchedoligosaccharides wasanalyzedbymoleculardocking calculations.Toaccomplish this, seven branched oligosaccharides were selected based on previouslyreporteddataonalgallectins,whichsharethesame mannopentosecore,varyingbyextramannoseresiduesat differ-entbranches(Fig.1)[7,12].Thecarbohydratestructureswerebuilt throughthewebserverSWEETII[34,35].
CalculationswereperformedwithAutoDockVina,version1.1.2, whichappliesaniteratedlocalsearchglobaloptimizerforthe opti-mizationprocedure,wherethesuccessionofeachstepconsistsof amutationandlocaloptimization,withtheacceptancedecisions made according to the Metropolis criterion. It uses the effi-cientquasi-NewtonmethodBroyden–Fletcher–Goldfarb–Shanno (BFGS)forlocaloptimization[36].TheAutodockgraphicalinterface AutoDockTools,version1.5.6,wasusedtokeeppolarhydrogens and add partial charges to the proteins and ligands using the KollmanUnitedcharges[37].Theproteinand carbohydrate lig-andsweretreatedasrigidandflexiblemolecules,respectively.The searchspaceforthedockingcalculations,selectedbasedonthe lengthofmucinfragments,wasdefinedbya20Å×20Å×20Åcube
centredontheconservedcarbohydratebindingsites. Exhaustive-nesswassetto15,andallotherparameterswereusedasdefault. Foreachdocking,thetentop-rankedgenerationsbasedonthe pre-dictedbindingaffinity(inkilocalories permole)wereanalyzed. Thesolutionswerefirstchosenbasedonthecoordinationofthe mannopentosecoreco-crystallizedwithOAAandBOA.Themost suitableresultswerefurtherrankedbasedonthetheoretical bind-ingenergy,whichisgivenasanegativescoreinkcal/mol.
theroot-mean-squaredeviation(RMSD)ofheavyatompositions betweenthedockedconformationandthecrystalstructurewas usedtoassesstheaccuracyofdockingpredictions.Basedon previ-ouslyreportedanalysis,wedefinedasolutionthathadamaximum RMSDvalueof2.0Åasanacceptabledockedresult[38,39].RMSD analysiswasperformedwiththeVisualMolecularDynamics(VMD) software[40].
2.6. Anticancereffect
2.6.1. Celllineandcultureconditions
Humanbreastadenocarcinomacellline(MCF-7)andprimary Human Dermal Fibroblasts (HDF) were both purchased from American Type Culture Collection (ATCC, USA). The cells were maintainedinT-25flaskscontainingDulbecco’smodifiedEagle’s medium (DMEM; Gibco®, TX, USA) supplemented with 10% of heat-inactivatedFetalBovineSerum(FBS;Gibco®
,TX,USA),1%ofl
-glutamine,100UmL−1penicillinand100gmL−1streptomycinat
37◦Cinahumidifiedatmospherecontaining5%CO2.Mediumwas routinelychangedevery3rdday,andcellsat90%confluencewere subculturedbytrypsinization(0.025%trypsin/0.1% EDTA).Inall experiments,cellswereusedbetweenthe3rdand14thpassage.
2.6.2. Cellviabilityassay
CellviabilitywasdeterminedusingCellTiter96®
AqueousMTS ReagentPowder(Promega,WI,USA),followingthemanufacturer´ıs instructions.Briefly,cells(2×104/200L/well)wereseededinto a96-well flatbottom microtiterplateinDMEMcontaining10% FBS and incubated overnight.Afterwards, the supernatant was removedandreplacedbyDMEMsupplementedwith2%FBS con-tainingdifferentconcentrationsofSfL.MTSassaywasperformed during24hforMCF-7cellsandHDF.
Fourindependentexperiments wereperformed intriplicate. Theopticaldensitywasreadat490nmonamicroplatereader.The viabilitywascalculatedas:
Cellviability (%)= averageOD490nm (control)averageOD490nm (H3) ∗100
2.6.3. AnnexinVassay
CellapoptosisandnecrosiswereassessedwiththeAPCAnnexin V ApoptosisDetectionKitwith7-AADdouble staining method. Briefly,MCF-7cells(2.25×105/well)wereseededin6-wellplates
containingDMEMsupplementedwith10%(v/v)FBS andgrown for 24h. Then, cells wereharvested and incubated withSfL at 125gmL−1 inDMEM containing2%FBSfor 24h. Controlcells wereincubatedwithDMEMsupplementedwith2%FBSonly.After treatmentwithSfLintheaforementionedperiods,thecellswere detachedbytrypsinizationandthencentrifuged(206g/5min)and washedtwicewithphosphatebufferedsaline(PBS;pH7.4).The cells were resuspended in binding buffer (PBS,pH 7.4, 25mM CaCl2),and5LofannexinV-APC(ImunoTools,Germany)and5L
of7AAD(InvitrogenTM,CA,USA)wereaddedtoeach well.Cells
wereincubatedinthedarkfor20minatroomtemperatureand thenthepro-apoptoticpotentialofSfLwasdeterminedbyflow cytometry(FCM).
2.6.4. mRNAextractionandqRT-PCR
Thequantificationofrelativegeneexpressionwasperformed byquantitativereversetranscriptase-realtimepolymerasechain reaction(qRT-PCR)aspreviouslydescribed[41].Briefly,after24h of treatmentwithSfL, MCF-7(106 cells) were trypsinized,and
totalmRNAwasextractedusingGenEluteMammaliantotalRNA Purificationkit(SigmaAldrich,MO, USA),andDNAse treatment wasperformedtoeliminateDNA(Nzytech,Lisboa,POR).Then,1g oftotal mRNAwasreversetranscribedusing theHighCapacity
cDNA Reverse Transcription kit (Applied Biosystems, CA,USA). qRT-PCRwasperformedina 7500FastSystem(Applied Biosys-tems, CA, USA), using TaqMan Fast Universal PCR Master Mix. PrimersandTaqManprobeswereacquiredfromApplied Biosys-tems and used following the manufacturer´ıs instructions. The analyzedgeneswereBAX(Hs00180269m1),BCL-2(Hs00608023), CASP 3(Hs00234387m1), CASP 8(Hs01018151m1), CASP 9(Hs00609647m1), and TP53(Hs01034249m1). Each reac-tionwasperformedinduplicate.Allgenes,includingendogenous controls (-actin and GAPDH) and treated or untreated cells, werealwaysanalyzedin thesame runtoexcludebetween-run variations.
Therelative expression for eachgene wascalculated by the 2−Ctmethodaspreviouslyreported[42].Theamplification
effi-ciencyforeachprimer/probewasabove95%.
2.6.5. Statisticalanalysis
Allresultswereconfirmedbyatleastthreeindependent exper-iments.Statisticsarepresentedasthemean±SEM.Experimental
datawereanalyzedbyStudent´ıst-testandone-wayANOVA fol-lowedbyTukey´ıspost-hoc.P<0.01andP<0.05wereadoptedas thelevelofsignificance.TheIC50valueswerecalculatedbyusing thesoftwareGraphPadPrism®
5tofitavariable sigmoidal-dose-responsecurve.
3. Results
3.1. PurificationSfL
Thelectinwaspurifiedbyacombinationof(NH4)2SO4
precipita-tionandion-exchangechromatographyonDEAE-SephacelColumn, asdescribedbyBenevidesandcollaborators[16].Apureproteinof relativemolecularmassof28kDawasobservedinSDSPAGE12.5% (datanotshown).
3.2. N-terminalaminoacidsequence
ThefirstfifteenaminoacidsofSfLweredeterminedbyEdman degradation.Heterogeneitieswereobservedinpositions4and5, indicatingtheexistenceofisoforms.ThrandAsnwereobserved in position 4, whereas Ala and Val were observed in position 5.Theresidue inposition13couldnotbeidentified.Therefore, theN-terminal ofSfL wasGRY(T/N)(A/V)QNQWGGSXAP.Search forsimilarityintheNCBIPROTEINBLASTshowedhighsimilarity betweenSfLandESA-2(P84331.1).
3.3. MolecularmassandaminoacidsequencingbyMS/MS
LC–MSshowedadistinctionseriesrelatedtoSfL(Fig.2).The majorionof27,553Daagreesverywellwiththeapparent molecu-larmassof28kDaobservedinSDS-PAGE.Othermolecularmasses wereobservedindeconvolutedspectra,indicatingthepresenceof isoformsinSfLpreparations.Smallvariationsaroundthese molec-ular masses were observed, suggesting the presence of adduct formations.
Fig.2. MolecularmassdeterminationofSfL.ESI–MSdeconvolutedmassspectraofLC–MSofSfLs.Insert.Zoomofregionofmassbetween27,900and28,100onthe deconvolutedmassspectra.
Fig.3.AminoacidsequencesofSfL-1(A)andSfL-2(B).Primarystructureswere determinedbycDNAcloningsequence,N-terminaldeterminationandTandemmass spectrometryoftrypticpeptides(T).Molecularmassofpeptides,asdeterminedby massspectrometry,isinparentheses.
3.4. PrimarystructuredeterminationofSfL-1andSfL-2
ThesequencingofthePCR3′RACEproductrevealedapartial aminoacidsequencefrom5to268,indicatingtwoisolectins,SfL-1 andSfL-2.TheprimarystructureofSfL-1andSfL-2wasdetermined byoverlap of sequencesobtainedby cDNA cloning,N-terminal determinationbyEdmandegradationandpeptidessequencedby MS/MS(Fig.3).
Thecalculated molecularmasses of thededucedaminoacid sequencesofSfL-1andSfL-2were27,552Daand27,985Da, respec-tively,whichwereconsistentwithions27,553Daand27,988Da, asdeterminedbyESI–MS.
The primary structure of SfL-1 and SfL-2 has four tandem-repeatdomains,consistingof67aminoacidseach.InSfL-1,the domainsshareatleast53.7%ofidentityeachother,whereasin SfL-2,identityamongdomainswasatleast44.7%(Fig.4).SfLisoforms sharedsequencesimilaritywithseverallectins(Fig.5),including OAA(P84330),PFL(WP016985694),BOA(WP010112459),MBHA
Table1
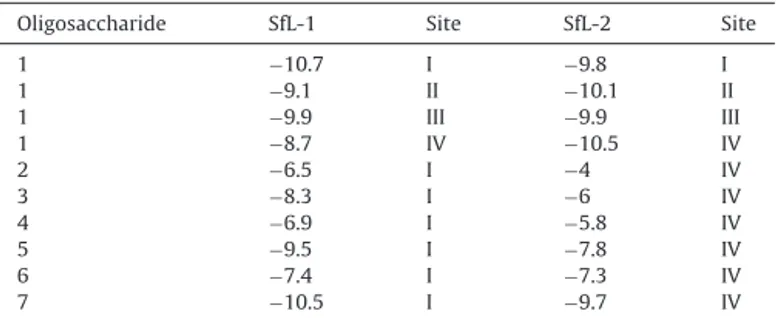
Theoreticalbindingenergy(kcal/mol)basedondockingcalculations.
Oligosaccharide SfL-1 Site SfL-2 Site
1 −10.7 I −9.8 I
1 −9.1 II −10.1 II
1 −9.9 III −9.9 III
1 −8.7 IV −10.5 IV
2 −6.5 I −4 IV
3 −8.3 I −6 IV
4 −6.9 I −5.8 IV
5 −9.5 I −7.8 IV
6 −7.4 I −7.3 IV
7 −10.5 I −9.7 IV
(WP 011556980),ESA-2(P84331),EDA-2(BAR91516)andKSA-2 (BAR91206).
3.5. Structuralmodeling
ThestructureofB.oklahomensisagglutinin(BOA,PDBID:4GK9) presentedthehighestrankingstructuralsimilaritytoSfL-1,withan identityof63%,accordingtoPSI-BLASTsearchfortemplateproteins onPDB(63%identity,74%positivity,and1%gaps).Therefore,the three-dimensionalstructureofSfLisoformsbuiltthrough compar-ativemodelingusingBOAstructureastemplatewasdeterminedto besuitableforstructuralandbindinganalysis(Fig.6).Theamino acidsequenceofBOAcrystalstructurecoversthefulllengthofSfL isoforms,exceptforthefirstfiveaminoacidsintheNterminus.
TheRamachandranplotforSfL-1showedthat100%ofresidues wereplacedinallowedregionswith98.11%onthefavoredzone. ForSfL-2,99.7%ofresidueswereplacedinallowedregionswith 98.11%onthefavoredzone,aswell.Asareference,thetemplate crystalstructureofBOAhad100%and98.0%residuesinthefavored andallowedzones,respectively.ThesedataindicatethatbothSfL modelledstructures,SfL-1andSfL-2,shouldbestableandreliable foranalyzingSfL-oligosaccharidebindingmodesthrough molecu-lardocking.
3.6. Docking
Dockingsystem(AutodockVina)suitabilityforcomplexligands, suchasoligosaccharides,wasanalyzedbasedonaredocking calcu-lationinvolvingthecrystalstructureofthecomplexbetweenBOA and␣3-␣6-mannopentaose[9].ThecalculatedRMSDbetweenthe bestdockingresultandthecrystalstructurewas1.3,whichattests tothereliabilityoftheadoptedsystemforthecurrentpurpose, despitetheconsiderableamountofrotatablebondspresentinthe usedoligosaccharides.
Fig.4.AlignmentoftherepeateddomainsofSfL-1(A)andSfL-2(B).Blackandwhiteboxesrepresentidenticalandnon-identicalaminoacids,respectively.
Fig.5. AlignmentofSfL-1andSfL-2andotherOAAH-familymembers.TheaminoacidsequencesofSfL-1and-2werealignedwithEucheumaserraagglutinin(ESA-2), Kappaphycusstriatusagglutinin(KSA-2),Eucheumadenticulatumagglutinin(EDA-2),Myxococcusxanthusagglutinin(MBHA),BurkholderiaoklahomensisEO147agglutinin (BOA),Oscillatoriaagardhiiagglutinin(OAA)andPseudomonasfluorescensPf0-1lectin(PFL).ResiduesunderlinedareaminoacidsofthebindingsiteaccordingtoWhitley etal.[9].Blackandwhiteboxesrepresentidenticalandnon-identicalaminoacids,respectively.
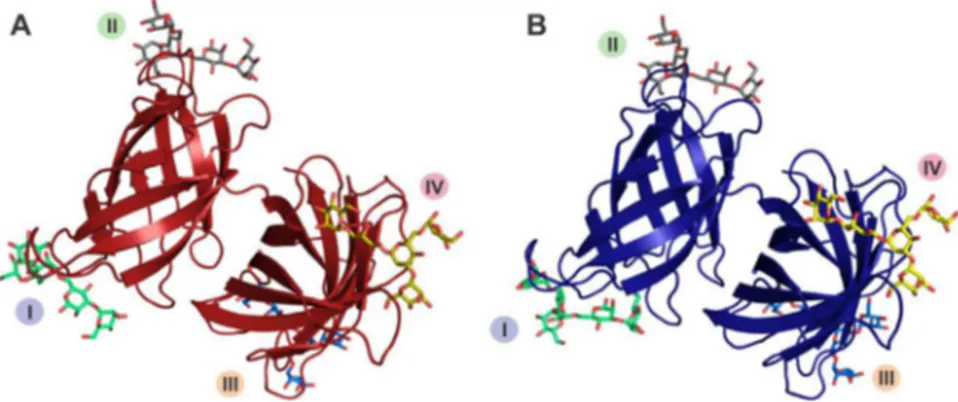
Fig.6. TheoverallstructureofSfL-1(A)andSfL-2(B).I,II,IIIandIVrepresentthecarbohydrate-bindingsites.
theothersixligands.For SfL-1,site Ipresentedthebestresult, whilesiteIVwasselectedforSfL-2.Despitethepresenceof con-servedresidues,subtledifferences inthecompositionof amino acidsdirectlyinvolvedinligandcoordinationamongthesitesin eachisoformandbetweenthem,beyondavariableloop structura-tion,mayinfluencethebindingspecificityofeachsite(Fig.7).
3.7. EffectofSfLonMCF-7cellviabilityandcellapoptosis
ToassesstheeffectofSfLoncellviability,weincubatedHDFand MCF-7cellswithincreasingdosesofthelectinat24h.Asshownin Fig.8,SfLdecreasedtheviabilityofMCF-7cells.Significantlosses of43.40%,58.27%and61.20%inviabilitywereobservedatdoses of62.5,250and500g.mL−1,respectively(*P<0.05).However,in
HDFcells,treatmentwithSfLdidnotcausereductionofmorethan
50%ofthecellpopulation.TheviabilityofHDFcellstreatedwith SfLat125and250gmL−1wasdecreasedbyonly34%and40.2%, respectively.At500gmL−1,aslightincreaseinthepopulation offibroblastswasobserved,suggestingthatthelectincan stimu-latecytoproliferationinlargerdoses.Afterwards,theconcentration that promoted50% inhibition of tumorcell viability(IC50)was
established,revealingthattreatmentwith125gmL−1 resulted
in50%inhibitionofMCF-7viability.
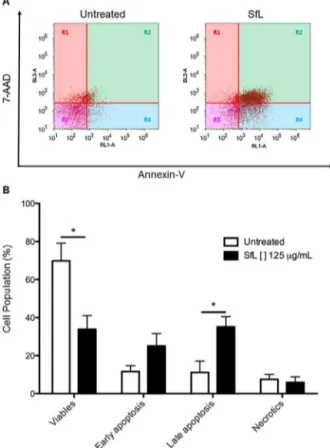
InordertoevaluatetheapoptoticpotentialofSfL,MCF-7cells weredoublestainedwithannexinV-APC/7AAD.Twenty-fourhours aftertreatmentwithSfLat125gmL−1,resultsshowedthat33.87%
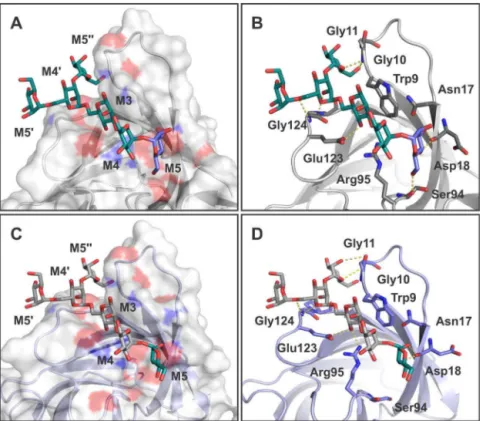
Fig.7. Analysisofligand7bindingtositeIonSFL-1andSFL-2.AandC–SurfacerepresentationofSfL-1andSfL-2,respectively,highlightingthebindingsitegrooveand ligandstructuration.BandD–Residuesinvolvedinthecoordinationofligand7onSfL-1andSfL-2(siteI),respectively,arerepresentedassticks,whilehydrogenbondsare presentedasdashedlines.Ligandsarerepresentedassticks.(Forinterpretationofthereferencestocolorinthisfigurelegend,thereaderisreferredtothewebversionof thisarticle.)
Fig.8. EffectofSfLonproliferationandcellviability.Humandermalfibroblasts andMCF-7cellsweretreatedwithserialconcentrationsofSfLduring24h.The cellviability(%)wasmeasuredbyMTSassay.(n=3,Mean±SEM).(*P<0.05and** P<0.01).
3.8. EffectofSfLonpro-apoptoticgeneexpression
TheeffectofSfLontheexpressionofpro-andanti-apoptotic genesofMCF-7cellswasanalyzed24haftertreatment.The rel-ativeexpressionofCASP3,CASP8,CASP9,BCL-2,BAX,andTP53 wasevaluatedbyreal-timePCR.MCF-7cells24haftertreatment withSfLwerecomparedwithuntreatedcells.AsshowninFig.10, after24h,theanti-apoptoticgeneBcl-2wasdownexpressed. How-ever,pro-apoptoticgenes,suchasBax,underwentover-expression. Interestingly,theexpressionofCASP3,-8and-9,whichareinvolved inintrinsicandextrinsicapoptosispathways,wereoverexpressed at24h(P=0.0139).These resultsindicatethat Solieriafiliformis
lectininducedcaspase-dependentapoptosiswherecaspase-8and -9arestronglyactivated.
4. Discussion
Inthecurrentstudy,wehaveisolatedanddeterminedthe pri-marystructureoftwoisolectinsfromS.filiformis,SfL-1andSfL-2. Weperformedastructuralpredictionforbothsequencesand ana-lyzedtheiranticancereffectonMCF-7breastcancercells.
Thedeterminedsequencesofbothisoforms,SfL-1andSfL-2, presentfourtandem-repeatdomainswithapproximately67amino acids,thuspresentinghighsequenceidentitytootherlectinsfrom theOAAH-family.SfL-1showed83%,83%,82%,62%and62%of iden-titywithESA-2,KSA-2,EDA-2,MBHAandBOA,respectively.SfL-2 showed80%,79%,78%,60%and60%ofidentitywithESA-2,KSA-2, EDA-2,MBHAandBOA,respectively.SfL-1andSfL-2showed82% ofidentitywitheachother.
Moreover,structuralmodelingandthebindinganalysis demon-stratethatSfL-1andSfL-2exhibitstructuralfeaturesandbinding specificitysimilartothelectinsoftheOAAH-family,indicatingthat SfL-1and-2couldbegroupedintothislectinfamily.
SimilartoBOA,SfL-1andSfL-2presentedstructurescomposed oftwo-barrel-likedomainsformedbyfiveantiparallel-strands, whichareconnectedbyashortpeptidelinker[9].Thelectin’sability tospecificallybindto3␣,6␣mannopentoseoligosaccharideswas assessedbymoleculardocking calculations,andfavorable bind-ingenergiesweredetectedforbothlectinisoforms,SfL-1(siteI) andSfL-2(siteIV),suggestingthesamespecificitytohigh-mannose oligosaccharidesasthatpresentedbylectinsoftheOAAH-family. Theaminoacidcompositioninvolved inthecarbohydrate bind-ingofSfL-1(siteI)wasW9G10G11N17D18S94R95E123G124P125
I126.InSfL-2(siteIV),itwasW211G212G213N219P220D161 R162
Fig.9.SfLcausedapoptosisinductioninMCF-7cells.MCF-7cellswereincubated with125g/mlSfLfor24h.AnnexinV-APC/7-AADdoublestainingwasperformed andanalyzedbyflowcytometry.(A)Dotblotsshowstagesofapoptosis/necrosis. R1(necroticcells),R2(lateapoptosis),R3(viablecells)andR4(apoptotic).(B) SfLinducesapoptosisinMCF-7cellswhencomparedtountreatedcells.(n=4, Mean±SEM).(*P<0.05).
Fig.10.SfLupregulatesmRNAexpressionofpro-apoptoticgenes.MCF-7cellswere treatedwithSfLataconcentrationof125g/mlfor24h.ThemRNAwasextracted andconvertedintocDNA.AnalysisofthemRNAlevelrelativetountreatedcellswas determinedbyreal-timePCR,usingthe2−Ctmethod.(**P<0.01).
residuesW152G153G154,R236andE264G265P266areinvolvedinthe
structurationofcarbohydratebindinginsiteIVinBOA.Theseamino acidsarehighlyconservedbetweendomainsandprimary struc-turesoflectinsfromtheOAAH-family.TheyarepresentinSfL-1 andSfL-2,participatingdirectlyincarbohydratebinding,as sug-gestedbymoleculardockingcalculations,pointingoutthesame specificityforSfLandtheOAAH-family.
Thedifferenceintheoreticalbindingenergyamongthesites betweentheisoformsoccursasaresultofminorchangesinthe
compositionandstructureofeachsite.SitesIandIIIofSfL-1and SfL-2havethesameaminoacidcomposition;however,inSfL-2, siteIisnarrower.SitesIIandIVaresimilarinstructureand vol-ume,buttheypresentadifferentaminoacidcomposition,suchas S26andA86insiteIIofSfL-1andG26andP86insiteIIofSfL-2.On
theotherhand,siteIVpresentsD161forSfL-1andS161forSfL-2.
Forthesimulationsinvolvingtheremainingoligosaccharides, the terminal GlcNAc residues were removed in order to avoid previously detected steric hindrances. Differently, the addition ofterminalmannoseresiduesonMan-7,Man-8andMan-9was accompanied by a decreased binding energy. These resultsare corroboratedby previousdataobtained forhomologous lectins [7,9,11,43,44].
LectinsfromtheOAAH-familyareknowntopresentantiviral andanticanceractivities,asreportedintheliterature[7–9,43,45]. Regarding theanticancer effect,SfL presented significant activ-ity, inhibiting 50% of MCF-7 cell viability with a treatment of 125gmL−1. Recently, increasing interest has been shown in marine lectins becauseof their pro-apoptotic,cytotoxic and antiproliferation effects over differentcell lines [46–49]. These lectinspresenthighspecificityforcomplexcarbohydrates. Con-sequently, their interaction with malignant, as compared to non-malignantcells,isbetterinviewofthelargenumberof glyco-proteinreceptorsontumorcellmembranes[50,51].Thus,theeffect ofSfLonnon-malignantcellsindicatesagreaterselectivityofSfL forMCF-7cells.Thisisaveryimportantpoint,sincemany anti-cancerdrugsgenerallydonotdifferentiatebetweennormaland malignantcells[52].Thepresentdatacorroboratewithpreviously reportedresultsforsimilarlectins,whichshowactivitiesagainst othercancercelllines[8,45].
Sugaharaandcollaborators[45]verifiedthatESAinducedcell death against Colo201 (Human Colon Cancer) and HeLa cells (humancervixcancer)atconcentrationsabove1.2gmL−1.
Nev-ertheless,MCF-7cellsshowedrelativelyhightolerancetoESA.The deathofColo201cellswasinvestigatedbyDNAladderdetection andcaspase-3activity,indicatingthatESAisabletoinduce apo-ptosisincancercellsafter3days.Thepro-apoptoticpotentialof SfLoverMCF-7cellswasanalyzedafter24h.SfLinducedthedeath of60.23%ofthecellpopulation,betweenearlyandlate apopto-sis,althoughmostcellswerein lateapoptosis.Manyterrestrial lectinshavetheabilitytoprovokecelldeath[53].However,littleis knownabouthowmarinelectinsinducenecrosisorapoptosis.To addressthisquestion,quantitativePCRassayswereperformedto verifyifthemechanismunderlyingcelldeathinducedbySfLtakes placebyintrinsicorextrinsicpathways.TheeffectofSfLafter24h ontheexpressionofpro-andanti-apoptoticgenesinMCF-7cells showedacaspase-dependentmechanismrelyingonCASP-8and -9,whichareproteinsrelatedtobothapoptosispathways.These datacorroboratethereportthatlectinswhichspecificallybindto N-glycancarbohydratemoietiesoncancercellsmayactaspotential therapeuticagentsviaapoptosisinduction[51].
Satoetal.[8]verifiedthatPFLpresentedasignificanteffecton decreasingthecellviabilityofMKN28cells(HumanGastricCancer) post-treatmentof0.5M(approximately6.94gmL−1)orhigher by72h,whichwasaccompaniedbythelossofcelladhesion,inturn triggeringasignalingpathwaythatinducedanoikis-likecelldeath. Furthermore,treatmentwithlow doses(0.1–0.3uM)stimulated theproliferationofMKN28cells.
carbo-hydrateligands[8,45].Therefore,thatfactthatSfLisalsoableto specificallybindtohigh-mannoseoligosaccharidespresentinthe MCF-7cellsstronglysuggeststhatitcanpotentiallyexertantitumor activity.However,togainabetterunderstandingofthatpotential, weneedtoundertakeindependentstudiesofeachisoform.
Acknowledgments
ThisworkwassupportedbytheBrazilianagenciesCNPq (Con-selho Nacional de Desenvolvimento Científico e Tecnológico), FUNCAP(Fundac¸ãoCearensedeApoioaoDesenvolvimento Cientí-ficoeTecnológico)andFINEP(FinanciadoradeEstudoseProjetos). TheauthorsaregratefultoProfessorDavidMartinforhelpingwith textediting.A.H.S., C.S.N.,and E.H.T.aresenior investigators of CNPq.
References
[1]R.S.Singh,S.R.Thakur,P.Bansal,Algallectinsaspromisingbiomoleculesfor biomedicalresearch,Crit.Rev.Microbiol.41(1)(2015)77–88.
[2]H.P.Singh,Mushroomlectinsaspromisinganticancersubstances,Curr. ProteinPept.Sci.17(8)(2016)797–807.
[3]C.S.Nagano,H.Debray,K.S.Nascimento,V.P.T.Pinto,B.S.Cavada,S. Saker-Sampaio,W.R.L.Farias,A.H.Sampaio,J.J.Calvete,HCAandHMLisolated fromtheredmarinealgaeHypneacervicornisandHypneamusciformisdefinea novellectinfamily,ProteinSci.14(8)(2005)2167–2176.
[4]K.S.Nascimento,C.S.Nagano,E.V.Nunes,R.F.Rodrigues,G.V.Goersch,B.S. Cavada,J.J.Calvete,S.Saker-Sampaio,W.R.Farias,A.H.Sampaio,Isolationand characterizationofanewagglutininfromtheredmarinealgaHypnea cervicornis,J.AgardhBiochem.CellBiol.84(1)(2006)49–54.
[5]K.Hori,K.Matsubara,K.Miyasawa,Primarystructuresoftwohemaglutinins frommarineredalgaHypneajaponica,Biochim.Biophys.Acta1474(2)(2000) 226–236.
[6]L.M.Koharudin,S.Kollipara,C.Aiken,A.M.Gronenborn,Structuralinsights intotheanti-HIVactivityoftheOscillatoriaagardhiiagglutininhomologlectin family,J.Biol.Chem.287(40)(2012)33796–33811.
[7]T.Sato,K.Hori,Cloning,expression,andcharacterizationofanovelanti-HIV lectinfromtheculturedcyanobacteriumOscillatoriaagardhii,FishSci.75(3) (2009)743–753.
[8]Y.Sato,K.Morimoto,T.Kubo,K.Yanagihara,T.Seyama,High
mannose-bindingantivirallectinPFLfromPseudomonasfluorescensPf0-1 promotescelldeathofgastriccancercellMKN28viainteractionwith a2-integrin,PLoSOne7(9)(2012)e45922.
[9]M.J.Whitley,W.Furey,S.Kollipara,A.M.Gronenborn,Burkholderia oklahomensisagglutininisacanonicaltwo-domainOAA-familylectin: structures,carbohydratebindingandanti-HIVactivity,FEBSJ.280(9)(2013) 2056–2067.
[10]J.M.Romeo,B.Esmon,D.R.Zusman,Nucleotidesequenceofthemyxobacterial hemagglutiningenecontainsfourhomologousdomains,Proc.Natl.Acad.Sci. U.S.A.83(17)(1986)6332–6336.
[11]K.Hori,Y.Sato,K.Ito,Y.Fujiwara,Y.Iwamoto,H.Makino,A.Kawakubo,Strict specificityforhigh-mannosetypeN-glycansandprimarystructureofared algaEucheumaserralectin,Glycobiology17(5)(2007)479–491.
[12]L.D.Hung,M.Hirayama,B.M.Ly,K.Hori,Purification,primarystructure,and biologicalactivityofthehigh-mannoseN-glycan-specificlectinfrom cultivatedEucheumadenticulatum,J.Appl.Phycol.27(4)(2015)1657–1669. [13]L.D.Hung,M.Hirayama,B.M.Ly,K.Hori,Biologicalactivity,cDNAcloningand
primarystructureoflectinKSA-2fromthecultivatedredalgaKappaphycus striatum(Schmitz)DotyexSilva,Phytochem.Lett.14(2015)99–105. [14]M.Hirayama,H.Shibata,K.Imamura,T.Sakaguchi,K.Hori,High-mannose
specificlectinanditsrecombinantsfromacarrageenophytaKappaphycus alvareziirepresentapotentanti-HIVactivitythroughhigh-affinitybindingto theviralenvelopeglycoproteingp120,Mar.Biotechnol.18(1)(2016) 144–160.
[15]L.M.I.Koharudin,W.Furey,A.M.Gronenborn,Novelfoldandcarbohydrate specificityofthepotentanti-HIVcyanobacteriallectinfromOscillatoria agardhii,J.Biol.Chem.286(2)(2011)1588–1597.
[16]N.M.B.Benevides,A.M.Leite,A.L.P.Freitas,Atividadehemaglutinantedaalga vermelhaSolieriafiliformis,R.Bras.Fisiol.Veg.8(2)(1996)117–122. [17]T.M.Abreu,L.M.C.M.Silva,E.S.Vanderlei,C.M.deMelo,V.R.Pereira,N.M.B.
Benevides,CytokineproductioninducedbymarinealgaelectinsinBALB/c micesplenocytes,ProteinPept.Lett.19(9)(2012)975–981.
[18]T.M.Abreu,N.A.Ribeiro,H.V.Chaves,R.J.Jorge,M.M.Bezerra,H.S.Monteiro, I.M.Vasconcelos,E.F.Mota,N.M.Benevides,Antinociceptiveand
anti-inflammatoryactivitiesofthelectinfrommarineredalgaSolieria filiformis,PlantaMed.82(7)(2016)596–605.
[19]S.Sameni,M.P.Hande,PlumbagintriggersDNAdamageresponse,telomere dysfunctionandgenomeinstabilityofhumanbreastcancercells,Biomed. Pharmacother.82(2016)256–268.
[20]L.A.Torre,R.L.Siegel,E.M.Ward,A.Jemal,Globalcancerincidenceand mortalityratesandtrends–anupdate,cancerepidemiology,Cancer Epidemiol.Biomark.Prev.25(1)(2016)16–27.
[21]Y.M.Amen,Q.Zhu,M.S.Afifi,A.F.Halim,A.Ashour,K.Shimizu,Newcytotoxic lanostanoidtriterpenesfromGanodermalingzhi,Phytochem.Lett.17(2016) 64–70.
[22]A.M.Scott,J.P.Allison,J.D.Wolchok,Monoclonalantibodiesincancertherapy, CancerImmun.12(2012)14.
[23]M.Bradford,Anal.Biochem.72(1976)248–254.
[24]U.K.Laemmli,Cleavageofstructuralproteinsduringtheassemblyofthehead ofbacteriophageT4,Nature227(5259)(1970)680–685.
[25]A.Shevchenko,H.Tomas,J.Havlis,J.V.Olsen,M.Mann,In-geldigestionfor massspectrometriccharacterizationofproteinsandproteomes,Nat.Protoc.1 (6)(2006)2856–2860.
[26]B.Ewing,L.Hillier,M.C.Wendl,P.Green,Base-callingofautomatedsequencer tracesusingphred.I.Accuracyassessment,GenomeRes.8(3)(1998) 175–185.
[27]B.Ewing,P.Green,Base-callingofautomatedsequencertracesusingphred.II. Errorprobabilities,GenomeRes.8(3)(1998)186–194.
[28]D.Gordon,C.Abajian,P.Green,Consed:agraphicaltoolforsequence finishing,GenomeRes.8(3)(1998)195–202.
[29]F.Corpet,Multiplesequencealignmentwithhierarchicalclustering,Nucleic Acids16(22)(1988)10881–10890.
[30]P.Gouet,E.Courcelle,D.I.Stuart,F.Metoz,ESPript:multiplesequence alignmentsinPostScript,Bioinformatics15(4)(1999)305–308.
[31]B.Webb,A.Sali,ComparativeproteinstructuremodelingusingMODELLER, Curr.Protoc.Bioinform.(2014),47:5.6:5.6.1–5.6.32.
[32]M.Shen,A.Sali,Statisticalpotentialforassessmentandpredictionofprotein structures,ProteinSci.15(11)(2006)2507–2524.
[33]I.W.Davis,A.Leaver-Fay,V.B.Chen,J.N.Block,G.J.Kapral,X.Wang,L.W. Murray,W.B.Arendall,J.Snoeyink,J.S.Richardson,D.C.Richardson, MolProbity:all-atomcontactsandstructurevalidationforproteinsand nucleicacids,NucleicAcidsRes.35(2007)375–383.
[34]M.Frank,S.Schloissnig,Bioinformaticsandmolecularmodelingin glycobiology,CellMol.LifeSci.67(16)(2010)2749–2772.
[35]A.Bohne,E.Lang,C.W.vonderLieth,SWEET–WWW-basedrapid3D constructionofoligo-andpolysaccharides,Bioinformatics15(9)(1999) 767–768.
[36]O.Trott,A.J.Olson,AutoDockvina.improvingthespeedandaccuracyof dockingwithanewscoringfunction,efficientoptimization,and multithreading,J.Comput.Chem.31(2)(2010)455–461.
[37]G.Morris,R.Huey,AutoDock4andAutoDockTools4:automateddockingwith selectivereceptorflexibility,J.Comput.Chem.30(16)(2009)2785–2791. [38]B.Kramer,M.Rarey,T.Lengauer,EvaluationoftheFLEXXincremental
constructionalgorithmforprotein–liganddocking,Proteins37(2)(1999) 228–241.
[39]G.Jones,P.Willett,R.C.Glen,R.Leach,R.Taylor,Developmentandvalidation ofageneticalgorithmforflexibledocking,J.Mol.Biol.267(3)(1997)727–748. [40]J.C.Phillips,R.Braun,W.Wang,J.Gumbart,E.Tajkhorshid,E.Villa,C.Chipot,
R.D.Skeel,L.Kal,K.Schulten,ScalablemoleculardynamicswithNAMD,J. Comput.Chem.26(16)(2005)1781–1802.
[41]P.A.Videira,D.Ligeiro,M.Correia,H.Trindade,Geneexpressionanalysisin superficialbladdercancer:comparasionoftwosuitableendogenous referencegenes,Curr.Urol.1(2007)145–150.
[42]K.J.Livak,T.D.Schmittgen,Analysisofrelativegeneexpressiondatausing real-timequantitativePCRandthe2(-DeltaDeltaC(T))Method,Methods25 (4)(2001)402–408.
[43]Y.Sato,K.Morimoto,M.Hirayama,K.Hori,Highmannose-specificlectin (KAA-2)fromtheredalgaKappaphycusalvareziipotentlyinhibitsinfluenza virusinfectioninastrain-independentmanner,Biochem.Biophys.Res. Commun.405(2)(2011)291–296.
[44]L.D.Hung,Y.Sato,K.Hori,High-mannoseN-glycan-specificlectinfromthe redalgaKappaphycusstriatum(Carrageenophyte),Phytochemistry72(9) (2011)855–861.
[45]T.Sugahara,Y.Ohama,A.Fukuda,M.Hayashi,A.Kawakubo,K.Kato,The cytotoxiceffectofEucheumaserraagglutinin(ESA)oncancercellsandits applicationtomolecularprobefordrugdeliverysystemusinglipidvesicles, Cytotechnology36(1–3)(2001)93–99.
[46]C.S.Bah,E.F.Fang,T.B.Ng,S.Mros,M.McConnell,D.BekhitAel,Purification andcharacterizationofarhamnose-bindingchinooksalmonroelectinwith antiproliferativeactivitytowardtumorcellsandnitricoxide-inducingactivity towardmurinemacrophages,J.Agric.FoodChem.59(10)(2011)5720–5728. [47]R.Matsumoto,Y.Fujii,S.M.Kawsar,R.A.Kanaly,H.Yasumitsu,Y.Koide,I.
Hasan,C.Iwahara,Y.Ogawa,C.H.Im,S.Sugawara,M.Hosono,K.Nitta,J. Hamako,T.Matsui,Y.Ozeki,Cytotoxicityandglycan-bindingpropertiesofan 18kDalectinisolatedfromthemarinespongeHalichondriaokadai,Toxins (Basel)4(5)(2012)323–338.
[48]L.Rabelo,N.Monteiro,R.Serquiz,P.Santos,R.Oliveira,A.Oliveira,H.Rocha, A.H.Morais,A.Uchoa,E.Santos,Alactose-bindinglectinfromthemarine spongeCinachyrellaapion(Cal)inducescelldeathinhumancervical adenocarcinomacells,Mar.Drugs10(4)(2012)727–743.
[50]G.V.Faheina-Martins,A.L.daSilveira,M.V.Ramos,L.F.Marques-Santos,D.A. Araujo,Influenceoffetalbovineserumoncytotoxicandgenotoxiceffectsof lectinsinMCF-7cells,J.Biochem.Mol.Toxicol.25(5)(2011)290–296. [51]R.C.Cheung,J.H.Wong,W.Pan,Y.S.Chan,C.Yin,X.Dan,T.B.Ng,Marine
lectinsandtheirmedicinalapplications,Appl.Microbiol.Biotechnol.99(9) (2015)3755–3773.
[52]D.FerreiraRda,J.G.Ferreira,M.C.Silva,R.A.Silva-Lucca,R.Mentele,E.J. Paredes-Gamero,T.C.Bertolin,M.T.DosSantosCorreia,P.M.Paiva,A. Gustchina,A.Wlodawer,M.L.Oliva,Crystalstructureofcrataevatapiabark protein(CrataBL)anditseffectinhumanprostatecancercelllines,PLoSOne8 (6)(2013)e64426.