w ww . e l s e v i e r . c o m / l o c a t e / b j p
Original
article
Isolation
and
characterization
of
2-hydroxy-9,10-anthraquinone
from
Streptomyces
olivochromogenes
(ERINLG-261)
with
antimicrobial
and
antiproliferative
properties
Chandrasekar
Balachandran
a,b,∗,
Veeramuthu
Duraipandiyan
a,c,
Yuvaraj
Arun
d,
Balachandran
Sangeetha
e,
Nobuhiko
Emi
b,
Naif
Abdullah
Al-Dhabi
c,
Savarimuthu
Ignacimuthu
a,f,
Yoko
Inaguma
b,
Akinao
Okamoto
b,
Paramasivan
T.
Perumal
daDivisionofMicrobiologyandCancerBiology,EntomologyResearchInstitute,LoyolaCollege,Chennai,India bDepartmentofHematology,FujitaHealthUniversity,Toyoake,Aichi,Japan
cDepartmentofBotanyandMicrobiology,AddirriyahChairforEnvironmentalStudies,CollegeofScience,KingSaudUniversity,Riyadh,SaudiArabia dOrganicChemistryDivision,CSIR-CentralLeatherResearchInstitute,Chennai,India
eDepartmentofToxicology,AdvinusTherapeuticsLtd,Bangalore,India
fVisitingProfessorProgramme,DeanshipofScientificResearch,CollegeofScience,KingSaudUniversity,Riyadh,SaudiArabia
a
r
t
i
c
l
e
i
n
f
o
Articlehistory: Received28August2015 Accepted21December2015 Availableonline29January2016
Keywords:
Streptomycesolivochromogenes Antimicrobial
Cytotoxic Moleculardocking
2-Hydroxy-9,10-anthraquinone
a
b
s
t
r
a
c
t
CurrentlyStreptomycesisoneofthemostimportantantibioticproducingmicroorganismsagainst sev-eraldiseases.InthepresentstudyStreptomycesolivochromogenesERINLG-261wasisolatedfromthesoil samplesoftheMudumalaihills,WesternGhats,India.Morphological,physiological,biochemicaland 16SrRNAstudiesstronglysuggestedthatthisisolatebelongedtothegenusStreptomyces.ERINLG-261 showedgoodantimicrobialactivityagainstdifferentbacteriaandfungiinMicromonosporafermentation medium.Theactiveethylacetateextractwaspackedincolumnchromatographyoversilicagelwhich ledtotheisolationof2-hydroxy-9,10-anthraquinoneastheactiveprinciple.Theisolatedcompound showedgoodantimicrobialactivityagainsttestedbacteriaandfungiinminimuminhibitory concen-tration(MIC)andminimumbactericidalconcentration(MBC)studies.Thecompoundshowedmoderate
invitroantiproliferativeactivityagainstA549andCOLO320cells.Thecompoundwassubjectedto molec-ulardockingstudiesfortheinhibitionofTopoisomerase,TtgRandBeta-lactamaseenzymeswhichare targetsforantimicrobials.Dockingresultsofthecompoundshowedlowdockingenergywiththese enzymesindicatingitsusefulnessasantimicrobialagent.Thisisthefirstreportofantimicrobialand antiproliferativeactivityof2-hydroxy-9,10-anthraquinoneisolatedfromStreptomycesolivochromogenes
alongwithmoleculardockingstudies.
©2016SociedadeBrasileiradeFarmacognosia.PublishedbyElsevierEditoraLtda.Allrightsreserved.
Introduction
Actinomyceteshavebeenespeciallyusefultothe pharmaceu-ticalindustry fortheirseeminglyunlimitedcapacity toproduce secondarymetaboliteswithdiversechemicalstructuresand bio-logical activities. Actinomycetes are Gram-positive filamentous bacteria,characterizedbyacomplexmorphologicdifferentiation
cycle accompanied by the production of numerous
extracel-lular enzymes as well as many kinds of bioactive secondary
metabolites having great structural and functional diversity
∗ Correspondingauthor.
E-mail:balasang@fujita-hu.ac.jp(C.Balachandran).
(Williams et al., 1983).Actinomycetesare known producersof structurally diverse metabolites namely, -lactam antibiotics, thienamycin,macrolides,streptomycin, erythromycin, anthracy-clines,daunorubicin,doxorubicin,polyketides,rapamycin,FK-506, peptideantibiotics,virginiamycin,pristinamycin,aminoglycosides, gentamicin and kanamycin(Demain,1999).Secondary metabo-lites are potent antibiotics which have made Streptomyces the primaryantibioticproducingorganismsexploitedbythe pharma-ceuticalindustry(RajaandPrabakaran,2011).Streptomycesisone of themost major sources of antibioticproducing microorgan-ismsand back-boneforcuringimportantdiseases.Streptomyces
are known to be producers of many secondary metabolites
which have different biological activities such as antibacterial, antifungal,antiparasitic,antitumor,inflammatoryresponsesand
http://dx.doi.org/10.1016/j.bjp.2015.12.003
immunosuppressiveactions(Demain,1999;Sanghvietal.,2014; Rambabu et al., 2014; Balachandran et al., 2015). In recent times,Streptomyces hasbeencalledas antibioticstore roomor library.More than 23,000bioactivesecondary metabolites
pro-ducedby microorganismshave beenreported and over 10,000
of these compounds are produced by actinomycetes (Raja and Prabakaran, 2011). Among actinomycetes, around 7600 com-poundsareproducedbyStreptomycesspecies(RajaandPrabakaran, 2011). Aouiche et al. (2014) had reported saquayamycins iso-latedfromStreptomycesspp.PAL114whichshowedgoodactivity againstCandidaalbicansM3andBacillussubtilisATCC6633.Huang etal.(2015)reportedtheisolationofnewcompoundsandfour
knowncompounds wereisolatedfromthemarineStreptomyces
griseusRSH0407suchasbutylhomononactate,butylnonactate, 8-actyl homononactic acid, homononactic acids, nonactic acid,
homononactylnonactate,homononactylhomononactate.Among
thesesevencompoundsbutylhomononactateshowedgood cyto-toxicproperties againstHCT-8, A2780,BGC-823, BEL-7402,and A549cells.
Naturally occurring anthraquinones form the largest group of plant and microbial secondary metabolites. Anthraquinone derivativesare wellrecognizedas important biologicallyactive componentsfrom microbesand plants (Ankeet al., 1980).The anthraquinonestypeofcompoundsshowedactivityagainstcertain diseases including antifungal, antibacterial, anticancer, antioxi-dant,antiviral,anti-inflammatoryandantihumancytomegalovirus (Barnardetal.,1995;Agarwaletal.,2000;Iizukaetal.,2004;Chen etal.,2007;Ifesanetal.,2009).Inthepresentstudyantibacterial, cytotoxicand moleculardocking properties of 2-hydroxy-9,10-anthraquinone (1)isolated from Streptomyces olivochromogenes
(ERINLG-261)wereassessed.
Materialsandmethods
IsolationofStreptomycesolivochromogenes
Thesoilsampleswerecollectedfromthedepthof5–15cmat Mudumalaihills,Nilgiris,WesternGhatsofTamilNadu,India. Iso-lationofStreptomycesolivochromogeneswasperformedbyserial dilutionusingdilutionplatetechnique(Balachandranetal.,2014a).
Morphologicalandbiochemicalobservations
CulturalandmorphologicalfeaturesofERINLG-261were char-acterizedfollowingthedirectionsgivenbytheISP(Shirlingand Gottlieb,1966)andtheBergey’sManualofSystematicBacteriology. Culturalcharacteristicsofpureisolatesinvariousmedia(ISP1–7) wererecordedafterincubationat30◦Cfor7–14days.Theshapeof cell,Gram-stain,color,thepresenceofsporesandcolony morphol-ogywereassessedonsolidISPagarmedium.Biochemicalreactions, differenttemperatures,NaClconcentration,pHlevel,pigment pro-duction,enzymereactionandacidorgasproductionweredone followingthemethodsofBalachandranetal.(2012,2014a,b).
16SrRNAgeneamplification
Genomic DNA of ERINLG-261 was isolated using the
meth-ods of Hipura Streptomyces DNA spin kit-MB 527-20pr from
Hi-media.The 16S ribosomal RNA gene was amplified by PCR
method using primers 27f (51AGTTTGATCCTGGCTCAG31) and
1492r(51ACGGCTACCTTGTTACGACTT31).EachPCRmixturein a finalvolumeof20lcontained10mMTris–HCl(pH.8.3),50mM KCl,1.5mMMgCl2,200MofeachdNTP,10pmolofeachprimer,
50ng of genomic DNA and 1U of Taq DNA Polymerase (New
EnglandBiolabsInc).PCRamplificationwasdetectedby1%agarose
gelelectrophoresisandwasvisualizedbyultraviolet(UV) fluores-cenceafterethidiumbromidestaining.ThePCRproductobtained wassequencedbyanautomatedsequencer(GeneticAnalyser3130, AppliedBiosystem,andUSA).Thesameprimersasabovewereused forthispurpose.Thesequencewascomparedforsimilaritywiththe referencespeciesofbacteriacontainedingenomicdatabasebanks usingtheNCBIBLASTavailableathttp://www.ncbinlm-nih.gov/. Thepartial16S rRNAgene sequence of isolateERINLG-261 has beendepositedintheGenBankdatabaseunderaccessionnumber KF061091.Aphylogenetictreewasconstructedusingthe neighbor-joiningDNAdistancealgorithmusingsoftwareMEGA(version4.1) (Tamuraetal.,2007).
Primaryantimicrobialscreening
PrimaryantimicrobialactivitywasevaluatedonModified
Nutri-entGlucose Agar medium (MNGA)by thecross streakmethod
againstvariouspathogenicmicroorganisms(Balachandranetal., 2014b).
Mediaoptimization
Four fermentation mediawere used for mediaoptimization suchasStreptomycesmedia-1(tryptone:17g,peptone:3g,NaCl: 5g,K2HPO4: 1.25g, pH7 and H2O: 1000ml), NutrientGlucose media-2(glucose:10g,peptone:5g,yeastextract:3g,NaCl:3g, beefextract:3g,pH7andH2O:1000ml),Bennettmedia-3 (glu-cose:10g,peptone:2g,yeastextract:1g,maltextract:1g,pH7 andH2O:1000ml),Micromonosporamedia-4(glucose:10g,starch: 24g,peptone:3g,meatextract:3g,yeastextract:5g,CaCO3:4g, pH7andH2O: 1000ml).ActivecultureERINLG-261 was inocu-latedinthesefourfermentationmediaandincubatedfor0day,2nd day,4thday,6thday,8thday,10thdayand12thday.After incuba-tionsecondarymetaboliteswereextractedusingCHCl3,EtOAcand butanol(1:1v/v).Alltheextractswerecheckedforantimicrobial activityagainstbacteriaandfungi.
Extractionofsecondarymetabolites
CultureinoculateoftheisolateERINLG-261wastakenin500ml Erlenmeyerflaskscontaining150mlofmedia-4andincubatedat 30◦Cinashaker(200rpm)for10days.After10thdaytheculture brothwascentrifugedat8000×gfor20mintoremovethebiomass. EqualvolumeofCHCl3,EtOAcandbutanol(1:1v/v)wereadded andshakeninaseparatingfunnel.Theprocesswasrepeatedthrice andtheextractswerecombined.Theextractwasdriedover anhy-droussodiumsulphateanddistilledinarotaryevaporatorandthe redresidueobtainedwasfinallydriedinvacuum.Thesecondary metaboliteproductionwascontinuedupto20l.
Columnchromatography
TheactiveEtOAcextract(9g)wassubjectedtosilicagel col-umnchromatography(Acme’s100–200mesh)(columnsize-60cm length/2.5cm).Thecolumnwassuccessivelyelutedwithhexane, hexane:EtOAcmixtureswithincreasingpolarityandfinallywith
EtOAc:MeOH andMeOH (eachfractionbeing100ml).Based on
Table1
CulturecharacteristicsofStreptomycesolivochromogenes(ERINLG-261)indifferentmedia.
Medium Growth Substratemycelium Aerialmycelium Spores Pigmentcolor
ISP1 Good Goldenyellow Darkgray Present Red
ISP2 Good Goldenyellow Darkgray Present Red
ISP3 Good Yellow Gray Moderate Red
ISP4 Good Yellow Gray Present Lightred
ISP5 Good Yellow Gray Present Red
ISP6 Good Yellow Gray Present Lightred
ISP7 Poor Poor Gray Poor Moderate
MNGA Good Yellow Darkgray Present Lightred
ISP1–7:InternationalStreptomycesProject;MNGA:modifiednutrientglucoseagar;+:present.−:absent.
250mmwithinternaldiameterof6.0mmandwasfilledwith sil-icaparticlesof15mdiameterbondedwithoctadecylsilane(YMC packODSA(250×6.0mm),15m).Themobilephasewas com-posedofACNandaqueousHOAc(15:85,v/v);itwasisocratically elutedataflow-rateof3ml/minandinjectionvolumewas100l. Elutionwasmonitoredat254nmandpeakfractionwascollected accordingtotheelutionprofile.Thepurecompoundwasobtained asyellowcrystalfromMeOH(97.66%)(retentiontime14.008and elutiontime20min).
Microbialorganisms
ThefollowingGramnegativeandGrampositivebacteria, clin-ical isolates and fungi were used for the experiment. Seven Gramnegativebacteria:EnterobacteraerogenesMTCC111,Shigella
flexneriMTCC1457,Salmonellaparatyphi-B,Klebsiellapneumonia
MTCC109, Pseudomonas aeruginosa MTCC741, Proteus vulgaris
MTCC1771andSalmonellatyphimuriumMTCC1251;fourGram
positive bacteria: Bacillus subtilis MTCC441, Micrococcus luteus
MTCC 106, Staphylococcus aureus MTCC 96 and Staphylococcus
epidermidis MTCC 3615; seven clinical isolates (isolated from
patient’s urine samples): Escherichia coli (ESBL-3984, Extended SpectrumBetaLactamase),Escherichiacoli(ESBL-3904),Klebsiella
pneumoniae (ESBL-3971), Klebsiella pneumoniae (ESBL-75799),
Klebsiellapneumoniae (ESBL-3894),Klebsiella pneumoniae
(ESBL-3967) and Staphylococcus aureus (MRSA− methicillin resistant, clinicalpathogen).Thereferencecultureswereobtainedfromthe Instituteof MicrobialTechnology (IMTECH), Chandigarh,
India-160036;CandidaalbicansMTCC227,Malassesiapachydermatisand
AspergillusflavuswereobtainedfromtheDepartmentof
Microbiol-ogy,ChristianMedicalCollege,Vellore,TamilNadu,India.Bacterial inoculumswerepreparedbygrowingcellsinMuellerHintonbroth (MHB)(Himedia) for 24hat 37◦C. Thefilamentous fungiwere grownonSabourauddextroseagar(SDA)slantsat28◦Cfor10days andthesporeswerecollectedusingsteriledoubleddistilledwater andhomogenized.YeastwasgrownonSabourauddextrosebroth (SDB)at28◦Cfor48h.
Antimicrobialassay
Antibacterialandantifungalactivitieswerecarriedoutusing diskdiffusionmethod(Balachandranetal.,2013).Zonesof inhibi-tionwererecordedinmillimetersandtheexperimentwasrepeated thrice.
Minimuminhibitoryconcentration(MIC)
MICstudiesoftheisolatedcompoundwereperformed accord-ingtothestandardreferencemethodsforbacteria(Balachandran etal.,2014a,b),filamentousfungi(CLSI,2008),andyeasts(NCCLS, 1999,2002).Therequiredconcentrations(100,50,25,12.5,6.25 and3.125g/ml)ofthecompoundweredissolvedinDMSO.
Minimumbactericidalconcentration(MBC)
Freshlypreparedtubescontainingserialtwofolddilutionsof thecompoundin5mlofMHB(range,100,50,25,12.5,6.25and 3.13g/ml)wereinoculatedbeneaththesurfacewith5×105 to 1×106cellsin0.1mlofMHB,mixedbyflushingsandincubated withoutshakingoragitation.After20hofincubation,allbroths wereexaminedfor visualturbidityorgrowthof smallcolonies at thebottomof tubesand againvortexed.The tubeswere re-incubatedforafurther4handvortexedagainuntilalltubeswere foundtobewithoutvisualturbidity.TheMBCwasconsideredas thelowestconcentrationofisolatedcompoundwhichprevented growthandreducedtheinoculumby≥99.9%within24h, irrespec-tiveofcountsofsurvivorsathigherantibioticconcentrationsand
Table2
Physiologicaland biochemicalcharacteristicsof Streptomycesolivochromogenes (ERINLG-261).
Characteristics Results
Gramstaining Positive
Shapeandgrowth Filamentousaerialgrowth
Productionofdiffusiblepigment + Rangeoftemperatureforgrowth 25–37◦C
Optimumtemperature 30◦C
RangeofpHforgrowth 6–10
NormalpH 7
H2Sproduction −
Amylase +
Chitinase +
Protease +
Gelatinase −
Indoleproduction +
GrowthinthepresenceofNaCl 1–7%
Sugaranalysis
Glucose ++
Galactose +
Lactose −
Mannitol −
Sucrose ++
Xylose +
Rhamnose ++
Ribose +
Arabinose −
Mannose ++
Maltose ++
Starch ++
Standardantibiotics
Ciprofloxacin S
Gentamicin S
Ampicillin S
Cephaloridine S
Streptomycin S
Erythromycin S
Vencomycin S
Amikacin S
Penicillin S
Rifamycin S
Norfloxacin S
KF061091/ERINLG-261/ (Streptomyces ol...
FJ007405.1| Streptomyces scabiei
AB741446.1| Streptomyces olivochromog...
HE577950.1| Streptomyces sp. IMCr03 C...
JQ670769.1| Streptomyces sp. 075013 C...
EU360178.1| Streptomyces sp. 18(2008)...
JQ289352.1| Streptomyces sp. GCTTACCA...
JX312315.1| Streptomyces sp. MCR26 CA...
FJ532409.1| Streptomyces olivochromog...
AB184737.1| Streptomyces olivochromog...
EU603343.1| Streptomyces sp. MJM3509 ...
GU045529.1| Streptomyces sp. SXY43 AT...
HE617217.1| Streptomyces sp. CTTAACAC...
JN866626.1| Streptomyces sp. TGCAGTCG...
EU370088.1| Streptomyces sp. AGTCGAAC...
EU054368.1| Streptomyces sp. ACATGCAG...
HQ398415.1| Streptomyces sp. CO178 AC...
AB045868.1| Streptomyces fimbriatus G...
JF728875.1| Streptomyces chartreusis ...
HQ607437.1| Streptomyces flavovariabi...
FJ481059.1| Streptomyces chartreusis ...
FJ796459.1| Streptomyces pseudovenezu...
GQ924491.1| Streptomyces sp. GSENDO-0...
AJ399481.1| Streptomyces pseudovenezu...
JF439617.1| Streptomyces plumbiresist...
EU603348.1| Streptomyces sp. MJM4120 ...
FR692114.1| Streptomyces sp. BK189 TT...
AF112159 Streptomyces sp. EF-52 TCACG... 82
87 99 99 99
67 68
75 61
66
Fig.1. Phylogenetictreederivedfrom16SrRNAgenesequencesshowingtherelationshipbetweenStreptomycesolivochromogenes(ERINLG-261)andtheotherspecies belongingtothegenusStreptomycesconstructedusingtheneighbor-joiningmethod.Bootstrapvalueswereexpressedaspercentagesof1000replications.
thelowest concentrationofthecompoundinhibitingthevisual growthofthetestculturesontheagarplate.Forfungi,theplates wereincubatedfor48–72hat28◦Candforbacteriatheplateswere incubatedfor24hat37◦C(ChennakesavaRaoetal.,2014).
Cytotoxicproperties
A549lungadenocarcinomacancercelllineandCOLO320 can-cercelllinewereobtainedfromNationalInstituteofCellSciences, Pune. A549 cell line was maintained in complete tissue
cul-turemedium Dulbecco’sModified Eagle’sMedium(DMEM) and
COLO320cancercelllineRoswellParkMemorialInstitutemedium (RPMI)with10%FetalBovineSerumand2mMl-Glutamine,along
with antibiotics (about 100 International Unit/ml of penicillin, 100g/mlofstreptomycin)withthepHadjustedto7.2.The cyto-toxicitywasdeterminedaccordingtothemethod(SaravanaKumar etal.,2014)withsomechanges.Cells(5000cells/well)wereseeded in96wellplatescontainingmediumwithdifferentconcentrations suchas500, 400,300,200,and100g/ml.Thecellswere culti-vatedat37◦Cwith5%CO2and95%airin100%relativehumidity. Aftervariousdurationsofcultivation,thesolutioninthemedium wasremoved.Analiquotof100lofmediumcontaining1mg/ml of3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide wasloadedintheplate.Thecellswereculturedfor4handthenthe solutioninthemediumwasremoved.Analiquotof100lofDMSO wasaddedtotheplate,whichwasshakenuntilthecrystalswere dissolved.Thecytotoxicityagainstcancercellswasdeterminedby
measuringtheabsorbanceoftheconverteddyeat540nminan enzymelinkedimmunesorbantassayreader.Cytotoxicityofeach samplewasexpressedasthehalfmaximalinhibitoryconcentration (IC50)value.TheIC50valueistheconcentrationoftestsamplethat causes50%inhibitionofcellgrowth,averagedfromthreereplicate experiments.Thepercentageofgrowthinhibitionwascalculated usingthefollowingformula;inhibition(%)=A−B/A×100(A– Con-trolgroupandB–Treatedgroup).
Moleculardockingstudies
MoleculardockingstudiesweredoneusingtheAutoDockTools (ADT)version1.5.6andAutoDockversion4.2.5.1dockingprogram. DockingstudieswereperformedbyIntel®
corei5CPU(2.53GHz) withWindows7operatingsystem.
Proteinstructurepreparation
DockedreceptorstructuresofDNATopoisomeraseIV(PDBID: 4EMV),TtgR(PDBID:2UXO)andbetalactamase(PDBID:4NK3) were obtainedfrom theProtein Data Bank. The co-crystallized ligandin thereceptor crystalstructure wasremoved.Thenthe
polar hydrogen atoms were added, lower occupancy residue
structures were deleted, and any incomplete side chains were replacedusingtheADT.FurtherADTwasusedtoremove
crys-tal water; Gasteiger charges were added to each atom, and
distancebetweendonorandacceptoratomsthatformeda hydro-genbondwasdefinedas1.9 ˚Awithatoleranceof0.5 ˚A,andthe acceptor–hydrogen–donoranglewasnotlessthan120◦.The struc-tureswerethen savedinPDBQT fileformat,for furtherstudies inADT.
Ligandstructurepreparation
Ligand 2Dstructure was drawnusing ChemDraw Ultra12.0
(ChemOffice, 2010). Chem3D Ultra 12.0 was used to convert
2D structure into 3D and the energy was minimized using
Chloroform
Microorganism
Microorganism
Microorganism
Microorganism
A
B
C
D
30 25 20
10 5 0
B.s M.l
S.a S.e
S.f S.p
K.p P.a
P.v S.t
C.a A.f
M.p
B.s M.l
S.a S.e
S.f S.p
K.p P.a
P.v S.t
C.a A.f M.p
B.s M.l
S.a S.e
S.f S.p
K.p P.a
P.v S.t
C.a A.f
M.p
B.s M.l
S.a S.e S.f S.p
K.p P.a
P.v S.t
C.a A.f M.p
Control n-Butanol Ethyl acetate Chloroform
n-Butanol Ethyl acetate Chloroform 15
Zone of incubation
Zone of incubation
Zone of incubation
30 25 20
10 5 0 15
30 25 20
10 5 0 15
30 25 20
10 5 0 15
Zone of inhibition
Ethyl acetate n-Butanol Control
Chloroform Ethyl acetate n-Butanol Control
Chloroform Ethyl acetate n-Butanol Control
Chloroform Ethyl acetate n-Butanol Control
Control
n-Butanol Ethyl acetate Chloroform
Control
n-Butanol Ethyl acetate Chloroform
Control
semi-empiricalAM1method.MinimizedenergytominimumRMS gradientof0.100wassetineachiteration.Allstructuresweresaved asPDBfileformatforinputtoADT.Alltheligandstructureswere thensavedinPDBQTfileformat,tocarryoutdockinginADT.
Gridformation
Agridboxwithdimensionof40×40×40 ˚A3with0.375 ˚A spac-ingandcenteredon(x,y,z)14.789,29.446,7.080;0.856,34.778, 13.333and5.475,13.239,18.265wascreatedaroundthebinding siteofligandonDNATopoisomeraseIV(PDBID:4EMV),TtgR(PDB ID:2UXO)andbetalactamase(PDBID:4NK3),respectively,using ADT.Thecenteroftheboxwassetatligandcenterandgridenergy calculationswerecarriedout.
Dockingprotocol
FortheAutoDockdockingcalculation,defaultparameterswere
used and 50 docked conformations were generated for each
compound.Theenergycalculationsweredoneusinggenetic algo-rithms.TheoutputswereexportedtoPyMolforvisualinspectionof thebindingmodesandinteractionsofthecompoundswithamino acidresiduesintheactivesites(PyMOL,2010).
Statisticalanalysis
Antimicrobial and cytotoxic activities of 2-hydroxy-9,10-anthraquinone(1)werestatisticallyanalyzedbyDuncanmultiple rangetestatp=0.05withthehelpofSPSS11.5versionsoftware package.
Table3
Minimuminhibitoryconcentrationandminimumbactericidalconcentrationof 2-hydroxy-9,10-anthraquinonefromStreptomycesolivochromogenes(ERINLG-261) againstbacteriaandfungi.
Organism MIC(g/ml) MBC(g/ml) Streptomycin
Gramnegative
E.aerogenes 50 50 25
K.pneumoniae 50 50 25
P.vulgaris 12.5 25 6.25
P.aeruginosa 12.5 12.5 25
S.paratyphi-B 25 25 6.25
S.typhimurium 25 25 25
S.flexneri 12.5 12.5 6.25
Grampositive
B.subtilis >100 >100 12.5
M.luteus >100 >100 6.25
S.aureus >100 >100 6.25
S.epidermidis 50 100 –
Clinicalisolates
E.coli(ESBL-3984) 25 25 25
E.coli(ESBL-3904) 50 100 25
K.pneumoniae (ESBL-3971)
12.5 12.5 6.25
K.pneumoniae (ESBL-75799)
50 50 25
K.pneumoniae (ESBL-3894)
>100 >100 6.25
K.pneumoniae (ESBL-3967)
>100 >100 25
S.aureus(MRSA) 50 100 6.25
Fungi Ketoconazole
C.albicans 50 100 25
A.flavus >100 >100 12.5
M.pachydermatis 25 50 15
Streptomycin–standardantibacterialagent;ketoconazole–standardantifungal agent.
Resultsanddiscussion
Morphologyandbiochemicalstudies
We were isolated 25 strains from different soil samples of theMudumalaihills,Nilgiris,WesternGhatsofTamilNadu,India usinghumic acidvitamin agarmedium.Among the25 isolates ERINLG-261 strain showed good antimicrobial activity in pre-liminary screening. This strain was Gram-positive filamentous bacterium.Thecolorofthesubstratemyceliawasgoldenyellow (Table1).ERINLG-261showedgoodgrowthonmediumamended withsodiumchlorideupto7%;nogrowthwasseenat9%.The temperatureforgrowthrangedfrom25 to37◦Cwithoptimum of30◦CandthepHrangewas6–10withoptimalpHof7. Uti-lizationofvariouscarbonsourcesbyERINLG-261indicatedawide patternofcarbonsourceassimilation.Starch,maltose,mannose, rhamnose,sucroseandglucosesupportedthegrowthofthestrain. ERINLG-261showedsensitivityinalltestedantibiotics(Table2). Theculture,morphologicalcharacteristicsandantimicrobial activ-itiesofdifferentStreptomycesisolateshavebeenreportedbyseveral investigators(Oskayetal.,2004).
16SrRNAgeneamplification
TheresultofthesequencingofERINLG-261wasobtainedinthe formofroughelectrophoregrams.Thephylogenetictreeobtained byapplyingtheneighborjoiningmethodisillustratedinFig.1. Cul-turecharacteristicsand16SrRNAstudiesstronglysuggestedthat ourisolateERINLG-261belongedtothegenusStreptomyces.Studies onthemicrobialdiversityby16SrRNAgeneanalysisshowedthat agroupofhigh-GCGram-positivebacteria(actinomycetes)were dominantinthesoil(Urakawaetal.,1999).Theidentificationof iso-lateERINLG-261wasconfirmedasStreptomycesolivochromogenes
withhomologyof100%.
Antimicrobialactivityofextracts
Streptomycesolivochromogenes(ERINLG-261)wasgrownin dif-ferentfermentationmedia-1to-4andextractedwithCHCl3,EtOAc and butanol. Each extracts(fermentation media1–4) of CHCl3, EtOAcand butanol were testedagainst bacteria and fungi.The EtOAcextract(fermentationmedia-4)showedgoodantibacterial andantifungalactivitiesagainsttestedbacteriaandfungicompared toCHCl3andbutanolextracts(5mg/ml)(Fig.2).Secondary metabo-liteproductionwascheckedindifferentdaysofincubationswith
fermentationmedia-4. Maximumsecondarymetabolite
produc-tionwasobservedon10thdayanditshowedgoodantimicrobial activity(Fig.3).
Chloroform
100
90
80
Secondary metabolite production
70
60
50
40
30
20
10
0
0 2 4 6 8
Days
10 12 14
Ethyl acetate n-Butanol
Fig.3.Selectionofsecondarymetaboliteproductionusingfermentationmedia-4 (Micromonospora)atdifferentdaysofincubation.10thdayshowedgoodsecondary
Isolationofactiveprinciple
TheactiveprincipleobtainedbypreparativeHPLCfromfraction
6 as themajor compound gave yellowcrystals frommethanol
(50mg); it gave a blue color with alcoholic FeCl3 for phenol andpinkcolorwithalcoholicNaOH.On TLCoversilicagelwith
EtOAc:MeOH (9:1) as the developing system it gave a single
spot(Rf 0.39), yellowin color which onexposureto ammonia vapour turnedpink.The compound was C14H8O3 [M+H]+, m/z 225,onthebasisof1HNMRand13CNMR(dept)andMass.mp 301–302◦C[lit. 298–299◦C].UV:
maxMeOHnm:225, 249,278 and322.IR:
maxKBrcm−1:3413(hydroxyl),2952,2821,1671 (quinonecarbonyl), 1618,1570,1536, 1373,1331, 1256,1223, 1175,1148,1023,871, 815, 785(aromatic). 1HNMR (ı, CDCl3, 400MHz):8.35(2H,m,H-5andH-8),7.84(2H,m,H-6andH-7), 7.42(1H,d,H-1),6.82(1H,d,H-3),7.08(1H,s,H-4),5.14(1H,s, OH).13CNMR(␦,CDCl3,100MHz):181.2(C-9,C-10),152.5(C-3), 135.1 (C-6, C-7), 133.5 (C-8a,C-10a), 126.6 (C-9a, C-4a),131.2 (C-5,C-8),130.2(C-1),118.5(C-2),117.5(C-4).The1H and13CNMRshowedthecompoundtobe2-hydroxy-9,10-anthraquinone
(1). On the basis of the physical and spectroscopic data the
compound was identified as 2-hydroxy-9,10-anthraquinone.
Physical and spectroscopic data (UV, FT-IR, 1H NMR, 13C
NMR and MASS) were compared with literature (Saha et al.,
2013).
A
B
D
C
100
90
80
70
60
50
40
30
20
10
0
50 100 200 300 400 500
Concentration μg/ml
Cytoto
xicity
, %
Fig. 4. Cytotoxicity properties of 2-hydroxy-9,10-anthraquinone (1) against COLO320cells.Dataaremean ± SDofthreeindependentexperimentswitheach experimentconductedintriplicate.Positivecontrol-Cyclophosphamideata con-centrationof90±0.00156g/ml(IC50).
OH O
O
1
MICandMBCvaluesofisolatedcompound
The compound showed potent antibacterial and antifungal
activities.TheMIC andMBCvalues ofisolated compound were seenagainstE.aerogenes,S.flexneri,S.paratyphi-B,K.pneumoniae,
P.aeruginosa,P.vulgarisandS.typhimurium;someclinicalisolates wereE.coli(ESBL-3984),E.coli(ESBL-3904),K.pneumoniae (ESBL-3971), K.pneumoniae (ESBL-75799),S.aureus (MRSA)and fungi
M.pachydermatisandC.albicans(Table3).Maximumgrowthand pigmentproductionwereobservedinglucoseasthesolesource of carbon. Theoptimum temperature of30◦C wasfoundto be effectiveforgrowthandpigmentproduction.Maximum antimi-crobialcompoundwasobtainedatpH7.0.Earlierreportshowed thattwelveactinomycetesstrainswereisolatedfromthesoil sam-plesoftheHimalayasandERIH-44showedbothantibacterialand antifungalactivity(Duraipandiyanetal.,2010).Normallyantibiotic production was higher in medium having glucose (1%) as car-bonsource.Streptomycesolivochromogenes(ERINLG-261)showed
A
B
C
100
90
80
70
60
50
40
30
20
10
0
50 100 200 300 400 500
Concentration μg/ml
Cytoto
xicity
, %
goodantimicrobialactivityinMicromonosporamediumand indi-catedthattheantimicrobialcompoundswereextracellular.Most ofthe secondary metabolitesand antibioticswere extracellular in natureand extracellular productsof actinomycetesshowed potentantimicrobialactivities(Bernanetal.,1994;Haceneetal., 2000).Thestudyoftheinfluenceofdifferentnutritionalmediaand cultureconditions onantimicrobialcompound production indi-catedthat thehighest biologicalactivitieswere obtainedwhen
Micromonosporamediumwasusedasabase.Infact,ithasbeen shownthat thenatureof carbonand nitrogensources strongly affectedantibioticproductionindifferentorganismsandthe antibi-oticproductionwasincreasedbyglucoserichmedium(Cruzetal., 1999). P.aeruginosahas emerged asone of themost problem-aticGram-negativepathogen,withanalarminglyhighantibiotic resistancerate(Bacq-Calberget al., 1999).Evenwith themost effectiveantibioticsagainstthispathogen,namelycarbapenems (imipenemandmeropenem),theresistanceratewasfoundtobe 15–20.4%amongst152P.aeruginosastrains(Savafietal.,2005). Thepresentstudyshowedthattheisolatedcompoundwasactive againstP.aeruginosa.Thisactivitymightbeduetotheirabilityto complexwithbacterialcellwall(Cowan,1999)thus,inhibitingthe microbialgrowthandthemembranedisruptioncouldbesuggested asthemechanismofaction(Arvindetal.,2004).The antimicro-bialcompoundfromStreptomycesolivochromogenes(ERINLG-261) wasrecoveredusing ethyl acetate solvent.Mostof the antimi-crobialcompoundsareextractedusingethylacetate(Sosioetal., 2000).Moreover,threebioactivecompoundsof3-phenylpropionic acid, anthracene-9,10-quinone and 8-hydroxyquinoline showed strong antibacterial and antifungal activities (Narayana et al., 2008).Balachandranetal.(2014)hadreported 2,3-dihydroxy-9,10-anthraquinone isolated from Streptomyces galbus (ERINLG-127)
which showedgoodantimicrobialactivityagainst tested bacte-ria and fungi. Duraipandiyan et al. (2014) had reported novel 1,5,7-trihydroxy-3-hydroxymethylanthraquinoneisolatedfrom terrestrial Streptomyces sp. (ERI-26) which showed significant antimicrobial activity against Staphylococcus aureus, Staphylo-coccus epidermidis, Bacillus subtilis, Epidermophyton floccosum,
Aspergillusniger,Aspergillerflavus,TrichophytonrubrumandBotrytis cinerea.
Cytotoxicpropertiesofisolatedcompound
The isolated compound 2-hydroxy-9,10-anthraquinone (1) showedmoderatecytotoxicactivityinvitroagainstA549lungand COLO320cells.Itshowed62.7%activityatthedoseof500g/ml withIC50valueof400g/mlagainstCOLO320cells(Fig.4).Isolated compoundshowed54.7%cytotoxicityagainstA549cellsatthedose of500g/ml(Fig.5).Allconcentrationsusedintheexperiment decreasedthecellviabilitysignificantly(P<0.05)ina concentra-tiondependentmanner.Anumberofanthraquinoneshavebeen reportedtopossesstumorcellinhibitoryeffectsandarecurrently utilizedasclinicalanticanceragents.Anthraquinoneshavebeen shown toinhibit cancercells through a varietyof mechanisms includinginductionofapoptosis,intercalationand bindingwith cellularDNA, redox-cyclingradicalformation, and inhibition of topoisomerase(Pattersonetal.,1983;Fisheretal.,1990;Barasch etal.,1990;MuellerandStopper,1999;Leeetal.,2001;Lee,2001). Anew,highlyoxygenatedangucyclinonegephyromycinwas iso-latedfromanextractofaStreptomycesgriseusstrain.Gephyromycin exhibitedglutaminergicactivitytowardneuronalcells(Bringmann etal.,2005).
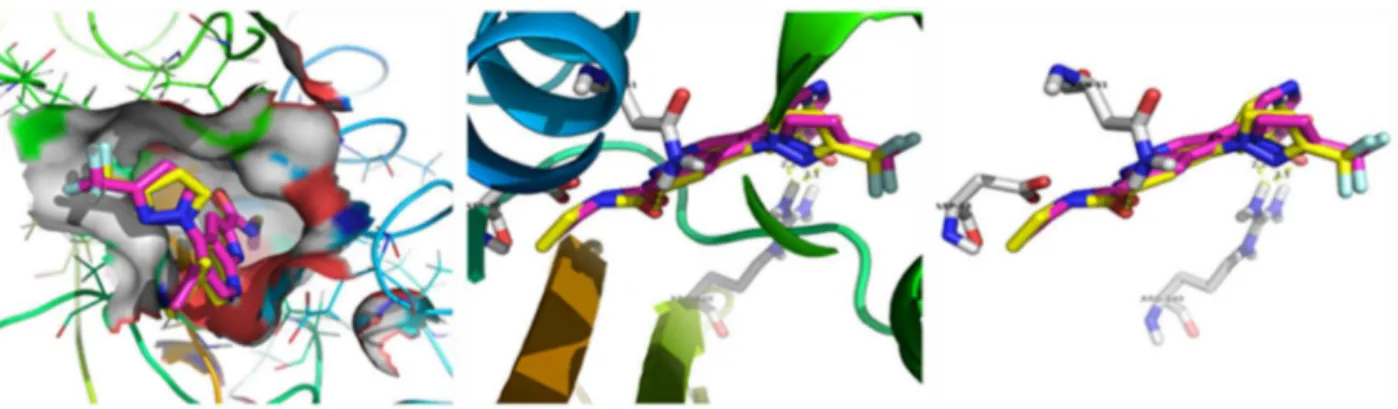
Method validation using crystallised and docked ligand of DNA Topoisomerase IV (4EMV)
A
B
Binding mode of compound with active site amino acids of DNA Topoisomerase IV (4EMV)
Moleculardockinganalysis
The compound was subjected to molecular docking studies
usingtheAutoDockTools(ADT)version1.5.6andAutoDockversion 4.2.5.1dockingprogram(Sanner,1999)toinvestigatethe poten-tialbindingmodeofinhibitor.Moleculardockingwasperformed withtheDNATopoisomerase IV(PDBID:4EMV),TtgR(PDBID: 2UXO)andbetalactamase(PDBID:4NK3)receptors.DNA topo-isomeraseIVreceptorisrequiredformaintenanceofproperDNA topologyduringtranscriptionandreplicationinbacteria.TtgRis importanttargetforantibioticdrugsbecauseantibioticresistance is a major problemin antimicrobialdrug synthesis. Onemajor mechanismthatunderliesantibioticresistanceinbacteriaisthe activeextrusionoftoxiccompoundsthroughthemembranebound effluxpumpsthatareoftenregulatedatthetranscriptionallevel. TtgR repressesthe transcription of TtgABC, a key effluxpump, whichishighlyresistanttoantibiotics(Manchesteretal.,2012). Beta-lactamskillbacteriabyinhibitingthecellwallconstruction enzymesknownasPBPs.However,somebacteriahavedeveloped enzymesthatcandestroybeta-lactamsbeforetheycaninactivate thePBPs.Theseenzymes,knownasbeta-lactamases,thusenable thebacteriatosurviveeveninthepresenceofhighconcentrations ofbeta-lactams.Hencetargetingthisbeta-lactaseenzymeisvital intheantibacterialdrugdesign(Algueletal.,2007).
In order to verify the reproducibility of the docking calcu-lations,thebound ligandwasextracted from thecomplex and submitted for one-ligand run calculation. This reproduced top scoringconformationfallingwithinroot-mean-squaredeviation (rmsd) value of 0.65 ˚A, 0.74 ˚A and 1.14 ˚A from bound X-ray
conformationof4EMV,2UXOand4NK3respectively,suggesting thatthismethodisvalidenoughtobeusedfordockingstudiesof othercompounds.
Docking of the compound to DNA Topoisomerase IV, TtgR
and beta lactamase was performed using AutoDock, following
thesame protocol used asin that of validation study. Docking was takeninto 2.5 million energy evaluations were performed forthetestmolecule.Dockedligandconformationwasanalyzed intermsofenergy,hydrogenbonding,andhydrophobic interac-tionbetweenligandandreceptor.Detailedanalysesoftheligand receptor interactions werecarried out, and final coordinatesof theligandandreceptorweresaved.PyMolsoftwarewasusedfor displayofthereceptorwiththeligandbindingsite.Fromthe dock-ingscores,thefreeenergyofbinding(FEB)ofthecompoundwas calculated.
Molecular docking of compound with DNA
Topoisome-rase IV (4EMV) receptor showed the binding energy value of
−7.04kcal/molwithtwohydrogenbonds.Inthecompound, hydro-genofO–HinteractswiththeC Ooxygenofaminoacid(ASP-78) andformsahydrogenbondwiththebondlengthof2.2 ˚A. Further-more,oxygenoftheoneofC OinteractswithNH2hydrogenof aminoacid(ASN-51)andformsahydrogenbondwiththebond lengthof2.2 ˚A(Fig.6).Thecompoundshowedthebindingenergy valueof−6.85kcal/molwithtwohydrogenbondswiththedocked TtgR(2UXO)receptor.Inthecompound,oxygenofO–Hinteracts withtheN–Hhydrogenofaminoacid(ASP-172)andformsa hydro-gen bondwith thebond lengthof 2.7 ˚A. Also, hydrogen of the O–HinteractswiththeC Ooxygenoftheaminoacid(PHE-168) andformsahydrogenbondwiththebondlengthof2.1 ˚A(Fig.7).
Method validation using crystallised and docked ligand of TtgR (2UXO)
A
B
Binding mode of compound with active site amino acids of TtgR (2UXO)
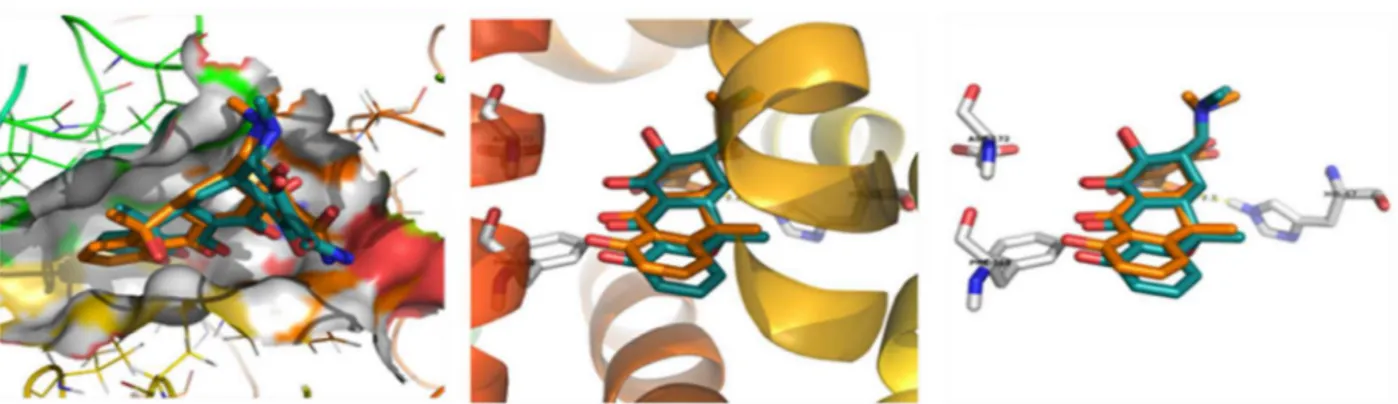
Method validation using crystallised and docked ligand of beta lactamase (4NK3)
A
B
Binding mode of compound with active site amino acids of beta lactamase (4NK3)
Fig.8.Putativebindingposeofcompound2-hydroxy-9,10-anthraquinonewithBeta-lactamase(4NK3).Dockingofcompound2-hydroxy-9,10-anthraquinonewith Beta-lactamase(A–methodvalidationandB–dockingwithaminoacids).
Thecompoundshowedthebindingenergyvalueof−6.26kcal/mol withtwohydrogenbondswiththedockedbetalactamase(4NK3) receptor.Inthecompound,oxygenofO–HinteractswiththeN–H hydrogenofaminoacid(ARG-175)andformsahydrogenbondwith thebondlengthof2.1 ˚A.Also,oxygenoftheC Ointeractswiththe NH2hydrogenofaminoacid(ASN-373)andformsahydrogenbond withthebondlengthof2.4 ˚A(Fig.8).
Conclusion
Streptomycesolivochromogenes(ERINLG-261)wasisolatedfrom
the soil samples of the Mudumalai hills, Nilgiris, Western
Ghats of Tamil Nadu, India. Ethyl acetate extract of ERINLG-261 showed significant antimicrobial activities against tested Grampositive and Gramnegative bacterialpathogens and fila-mentous fungalpathogens. The bioactivityguided fractionation of the ethyl acetate led to the isolation of 2-hydroxy-9,10-anthraquinone(1) as theactive principle. The 2-hydroxy-9,10-anthraquinonewassubjectedtoantimicrobialactivity:itshowed goodantimicrobialactivityagainsttestedbacteriaandfungi.The 2-hydroxy-9,10-anthraquinonewasalsotestedagainstCOLO320
and A549 lung adenocarcinoma cancer cells. The
2-hydroxy-9,10-anthraquinoneshowedmoderatecytotoxicpropertiesagainst testedcells.Moleculardockingstudiesofisolatedcompound
2-hydroxy-9,10-anthraquinone withenzyme Topoisomerase, TtgR
andBeta-lactamaseshowedlowbindingenergy.Thisisthefirst reportfortheantimicrobialandcytotoxicpropertiesof 2-hydroxy-9,10-anthraquinoneisolatedfromStreptomycesolivochromogenes
(ERINLG-261).
Authors’contributions
Conceivedanddesignedtheexperiments:CB,VD,YA,BS,NE, NAAD,SI,YI,AOandPTP.Performedtheexperiments:CB,VD,YA, andBS.Analyzedthedata:CB,VD,YA,BS,NE,NAAD,SI,YI,AOand PTP.Preparedthemanuscript,CB,VD,YA,BSandSI.Allauthors readandapprovedthefinalmanuscript.
Conflictsofinterest
Theauthorsdeclarenoconflictsofinterest.
Acknowledgement
Theauthorswouldliketoextendtheirsincereappreciationto theDeanshipofScientificResearchatKingSaudUniversityforits fundingofthisresearchthroughtheResearchGroupproject num-berRGP-VPP-213.
AppendixA. Supplementarydata
Supplementarydataassociatedwiththisarticlecanbefound,in theonlineversion,atdoi:10.1016/j.bjp.2015.12.003.
References
Agarwal, S.K., Singh, S.S., Verma, S., Kumar, S., 2000. Antifungal activity of anthraquinonederivativesfromRheumemodi.J.Ethnopharmacol.72,43–46. Alguel,Y.,Meng,C.,Terán,W.,Krell,T.,Ramos,J.L.,Gallegos,M.T.,Zhang,X.,2007.
Anke, H., Kolthoum, I., Zihner, H., Laatsch, H., 1980. Metabolic products of microorganisms185.TheanthraquinonesoftheAspergillusglaucusgroup.I. Occurrence,isolation,identificationandantimicrobialactivity.Arch.Microbiol. 126,223–230.
Aouiche,A.,Bijani,C.,Zitouni,A.,Mathieu,F.,Sabaou,N.,2014.Antimicrobial activ-ityofsaquayamycinsproducedbyStreptomycesspp.PAL114isolatedfroma Saharansoil.J.Med.Mycol.24,17–23.
Arvind,S.,Reg,F.C.,Enzo,A.P.,2004.Identificationofantimicrobialcomponents ofanethanolicextractoftheAustralianmedicinalplant,Eremophiladuttonii. Phytother.Res.18,615–618.
Bacq-Calberg,C.M.,Coyotte,J.,Hoet,P.,Nguyem-Disteeche,M.,1999.Microbiologie. DeBoeckandLarcier,Bruxelles,pp.338.
Balachandran,C.,Duraipandiyan1,V.,Emi,N.,Ignacimuthu,S.,2015.Antimicrobial andcytotoxicpropertiesofStreptomycessp(ERINLG-51)isolatedfromSouthern WesternGhats.SouthIndianJ.Biol.Sci.1,7–14.
Balachandran,C.,Sangeetha,B.,Duraipandiyan,V.,KarunaiRaj,M.,Ignacimuthu,S., Al-Dhabi,N.A.,Balakrishna,K.,Parthasarathy,K.,Arulmozhi,N.M.,ValanArasu, M.,2014a.AflavonoidisolatedfromStreptomycessp(ERINLG-4)induces apo-ptosisinhumanlungcancerA549cellsthroughp53andcytochromecrelease caspase-dependantpathway.Chem.Biol.Interact.224,24–35.
Balachandran, C., Arun, Y., Duraipandiyan, V., Ignacimuthu, S., Balakrishna, K., Al-Dhabi, N.A., 2014b. Antimicrobial and cytotoxicity properties of 2,3-dihydroxy-9,10-anthraquinoneisolatedfromStreptomycesgalbus (ERINLG-127).Appl.Biochem.Biotechnol.172,3513–3528.
Balachandran,C.,Duraipandiyan,V.,Balakrishna,K.,Sundaram,L.R.,Vijayakumar, A.,Ignacimuthu,S.,Al-Dhabi,N.A.,2013.Synthesisandmedicinalpropertiesof plantderivedvilangin.Environ.Chem.Lett.11,303–308.
Balachandran, C., Duraipandiyan, V., Balakrishna, K., Ignacimuthu, S., 2012. Petroleumand polycyclic aromatic hydrocarbons (PAHs) degradation and naphthalenemetabolisminStreptomycessp(ERI-CPDA-1)isolatedfromoil con-taminatedsoil.Bioresour.Technol.112,83–90.
Barasch,D.,Zipori,O.,Ringel,I.,Ginsburg,I.,Samuni,A.,Katzhendler,J.,1990.Novel anthraquinonederivativeswithredoxactivefunctionalgroupscapableof pro-ducingfreeradicalsbymetabolism:arefreeradicalsessentialforcytotoxicity? Eur.J.Med.Chem.34,597–615.
Barnard,D.L.,Fairbairn,D.W.,O’Neill,K.L.,Gage,T.L.,Sidwell,R.W.,1995. Antihu-mancytomegalovirusactivityandtoxicityofsulfonatedanthraquinonesand anthraquinonederivatives.AntiviralRes.28,317–321.
Bernan, V.S., Montenegro,D.A., Korshalla, J.D., Maiese, W.M., Steinberg,D.A., Greenstein,M.,1994.Bioxalomycinsnewantibioticsproducedbythemarine Streptomyces sp. LL-31F508: taxonomy and fermentation. J. Antibiot. 47, 1417–1424.
Bringmann,G.,Lang,G.,Hamm,M.A.,Gulder,T.A.M.,Dieter,A.,Bull,A.T.,Stach, J.E.M.,Kocher,N.,Muller,W.E.G.,Hans-Peter,F.,2005.Gephyromycin,thefirst bridgedangucyclinone,fromStreptomycesgriseusstrainNTK14.Phytochemistry 66,1366–1373.
Chen,S.H.,Lin,K.Y.,Chang,C.C.,Fang,C.L.,Lin,C.P.,2007.Aloeemodininduced apo-ptosisinhumangastriccarcinomacells.Food.Chem.Toxicol.45,2296–2303. ChennakesavaRao,K.,Arun,Y.,Easwaramoorthi,K.,Balachandran,C.,Prakasam,T.,
EswaraYuvaraj,T.,Perumal,P.T.,2014.Synthesis,antimicrobialandmolecular dockingstudiesofenantiomericallypureN-alkylatedb-aminoalcoholsfrom phenylpropanolamines.Bioorg.Med.Chem.Lett.24,3057–3063.
ClinicalandLaboratoryStandardsInstitute(CLSI),2008.ReferenceMethodfor BrothDilutionAntifungalSusceptibilityTestingofFilamentousFungi;Approved StandardSecond–EditionCLSIDocumentM38-A2.ClinicalandLaboratory StandardsInstitute,940,WestvalleyRoad,Suite1400,Wayne,Pennsylvania, 19087-1898USA,ISBN1-56238-668-9.
Cowan,M.M.,1999.Plantproductsasantimicrobialagents.Clin.Microbiol.Rev.12, 564–582.
Cruz,R.,Arias,M.E.,Soliveri,J.,1999.Nutritionalrequirementfortheproductionof PyrazoloisoquinolinoneantibioticsbyStreptomycesgriseocirneusNCIMB40447. Appl.Microbiol.Biotechnol.53,115–119.
Demain,A.L.,1999.Pharmaceuticallyactivesecondarymetabolitesof microorgan-isms.Appl.Microbiol.Biotechnol.52,455–463.
Duraipandiyan,V.,Sasi,A.H.,Islam,V.I.H.,Valanarasu,M.,Ignacimuthu,S.,2010. AntimicrobialpropertiesofactinomycetesfromthesoilofHimalaya.J.Mycol. Med.20,15–20.
Duraipandiyan,V.,AL-Dhabi,N.A.,Balachandran,C.,KarunaiRaj,M.,ValanArasu,M., Ignacimuthu,S.,2014.Novel1,5,7-trihydroxy-3-hydroxymethylanthraquinone isolatedfromterrestrialStreptomycessp(ERI-26)withantimicrobialand molec-ulardockingstudies.Appl.Biochem.Biotechnol.174,1784–1794.
Fisher,G.R.,Brown,J.R.,Patterson,L.H.,1990.Involvementofhydroxylradical for-mationandDNAstrandbreakageinthecytotoxicityofanthraquinoneantitumor agents.Free.Radic.Res.Commun.11,117–125.
Hacene,H.,Daoudi,H.,Bhatnagar,T.,Baratti,J.C.,Lefebvre,G.,2000.H107,anew aminoglycosidaseantiPseudomonasantibioticproducedbyanewstrainof Spir-illosora.Microbios102,69.
Huang,H.,Lan,X.,Wang,Y.,Tian,L.,Fang,Y.,Zhang,L.,Zhang,K.,Zheng,X.,2015. NewbioactivederivativesofnonacticacidfromthemarineStreptomycesgriseus derivedfromtheplantSalicorniasp.Phytochem.Lett.12,190–195.
Ifesan,B.O.,Hamtasin,C.,Mahabusarakam,W.,Voravuthikunchai,S.P.,2009. Assess-mentofantistaphylococcalactivityofpartially purifiedfractionsandpure compoundsfromEleutherineamericana.J.FoodProt.72,354–359.
Iizuka,A.,Iijima,O.T.,Kondo,K.,Itakura,H.,Yoshie,F.,Miyamoto,H.,Kubo,M., Higuchi,M.,Takeda,H.,Matsumiy,T.,2004.Evaluationofrhubarbusing antiox-idativeactivityasanindexofpharmacologicalusefulness.J.Ethnopharmacol. 91,89–94.
Lee,H.Z.,Hsu,S.L.,Liu,M.C.,Wu,C.H.,2001.Effectsandmechanismsofaloeemodin oncelldeathinhumanlungsquamouscellcarcinoma.Eur.J.Pharmacol.431, 287–295.
Lee,H.Z.,2001.ProteinkinaseCinvolvementinaloeemodinandemodininduced apoptosisinlungcarcinomacell.Br.J.Pharmacol.134,1093–1103.
Manchester,J.I.,Dussault,D.D.,Rose,J.A.,Boriack-Sjodin,P.A.,Uria-Nickelsen,M., Ioannidis,G.,Bist,S.,Fleming,P.,Hull,K.G.,2012.Discoveryofanovelazaindole classofantibacterialagentstargetingtheATPasedomainsofDNAgyraseand TopoisomeraseIV.Bioorg.Med.Chem.Lett.22,5150–5156.
Mueller, S.O., Stopper, H., 1999. Characterization of the genotoxicity of anthraquinonesinmammaliancells.Biochim.Biophys.Acta1428,406–414. Narayana,K.J.P.,Kumar,K.G.,Vijayalakshmi,M.,2008.l-Asparaginaseproduction
byStreptomycesalbidoflavus.IndianJ.Microbiol.48,331–336.
NationalCommitteeforClinicalLaboratoryStandards(NCCLS),1999.Document M31-APerformanceStandardsforAntimicrobialDiskandDilution Suscepti-bilityTestsforBacteriaIsolatedfromAnimals.NationalCommitteeforClinical LaboratoryStandards,Villanova,pp.57(approvedstandardNCCLS).
NationalCommitteeforClinicalLaboratoryStandards(NCCLS),2002.Reference MethodforBrothDilutionAntifungalSusceptibilityTestingofYeasts:Proposed Standard.NationalCommitteeforClinicalLaboratoryStandards(NCCLS), Vil-lanova.
Oskay,M.,Tamer,A.U.,Azeri,C.,2004.Antibacterialactivityofsomeactinomycetes isolatedfromfarmingsoilsofTurkey.Afr.J.Biotechnol.3,441–446.
Patterson, L.H., Gandecha,B.M., Brown,J.R., 1983. 1,4-Bis{2-[(2-hydroxyethyl) amino]ethylamino}-9,10-anthracenedione,ananthraquinoneantitumoragent thatdoesnotcauselipidperoxidationinvivo;comparisonwithdaunorubicin. Biochem.Biophys.Res.Commun.110,399–405.
Raja,A.,Prabakaran,P.,2011.Actinomycetesanddrug—anoverview.Am.J.Drug Dis.Dev.1,75–84.
Rambabu,V.,Suba,S.,Vijayakumar,S.,2014.Antimicrobialandantiproliferative prospectiveofkosinostatin–asecondarymetaboliteisolatedfromStreptomyces sp.J.Pharm.Anal.5,378–382.
Saha,K.,Lam,K.W.,Abas,F.,Hamzah,A.S.,Stanslas,J.,Hui,L.S.,Lajis,N.H.,2013. Synthesisofdamnacanthal,anaturallyoccurring9,10-anthraquinoneandits analogues,anditsbiologicalevaluationagainstfivecancercelllines.Med.Chem. Res.22,2093–2104.
Sanghvi,G.V.,Ghevariya,D.,Gosai,S.,Langa,R.,Dhaduk,N.,Kunjadia,P.D.,Vaishnav, D.J.,Dave,G.S.,2014.Isolationandpartialpurificationoferythromycinfrom alkaliphilicStreptomyceswerraensisisolatedfromRajkot,India.Biotechnol.Rep., 1–2,2-7.
Sanner,M.F.,1999.Python:aprogramminglanguageforsoftwareintegrationand development.J.Mol.Graph.Model.17,57–61.
SaravanaKumar,P.,Al-Dhabi,N.A.,Duraipandiyan,V.,Balachandran,C.,Praveen Kumar, P., Ignacimuthu, S., 2014. In vitro antimicrobial, antioxidant and cytotoxicpropertiesofStreptomyceslavendulaestrainSCA5.BMCMicrobiol., http://dx.doi.org/10.1186/s12866-014-0291-6.
Savafi,L.,Duran,N.,Savafi,N.,Onlem,Y.,Ocak,S.,2005.Theprevalenceand resis-tancepatternsofPseudomansaeruginosainintensivecareunitsinauniversity Hospital.J.Med.Sci.35,317.
Shirling,J.L.,Gottlieb,D.,1966.MethodsforcharacterizationofStreptomycesspecies. Int.J.Syst.Bacteriol.16,313–340.
Sosio,M.,Bossi,E.,Bianchi,A.,Donadio,S.,2000.Multiplepeptidesynthetasegene clustersinactinomycetes.Mol.Genet.Genomics264,213–221.
Tamura,K.,Dudley,J.,Nei,M.,Kumar,S.,2007.MEGA4:MolecularEvolutionary GeneticsAnalysis(MEGA)softwareversion4.0.Mol.Biol.Evol.24,1596–1599. Urakawa,H.,Kita-Tsukamoto,K.,Ohwada,K.,1999.Microbialdiversityinmarine sedimentsfromSagamiBayandTokyoBayJapan,asdeterminedby16SrRNA geneanalysis.Microbiology145,3305–3315.