CLINICAL SCIENCE
Antimicrobial resistance and prevalence of resistance
genes in intestinal
Bacteroidales
strains
Viviane Nakano,IAmanda do Nascimento e Silva,IVictor Rafael Castillo Merino,IHannah M. Wexler,II,III Mario Julio Avila-CamposI
IAnaerobe Laboratory, Department of Microbiology, Institute of Biomedical Sciences, Sa˜o Paulo University, Sa˜o Paulo, SP/Brazil.IIGreater Los Angeles
Veterans Administration Healthcare Systems, Los Angeles, CA, USA.IIIDepartment of Medicine, University of California, Los Angeles, CA, USA.
OBJECTIVE: This study examined the antimicrobial resistance profile and the prevalence of resistance genes in Bacteroidesspp. andParabacteroides distasonisstrains isolated from children’s intestinal microbiota.
METHODS:The susceptibility of these bacteria to 10 antimicrobials was determined using an agar dilution method.
b-lactamase activity was assessed by hydrolysis of the chromogenic cephalosporin of 114 Bacteriodales strains isolated from the fecal samples of 39 children, and the presence of resistance genes was tested using a PCR assay.
RESULTS: All strains were susceptible to imipenem and metronidazole. The following resistance rates were observed: amoxicillin (93%), amoxicillin/clavulanic acid (47.3%), ampicillin (96.4%), cephalexin (99%), cefoxitin (23%), penicillin (99%), clindamycin (34.2%) and tetracycline (53.5%).b-lactamase production was verified in 92% of the evaluated strains. The presence of thecfiA, cepA, ermF, tetQandnimgenes was observed in 62.3%, 76.3%, 27%, 79.8% and 7.8% of the strains, respectively.
CONCLUSIONS:Our results indicate an increase in the resistance to several antibiotics in intestinalBacteroidesspp. andParabacteroides distasonisand demonstrate that these microorganisms harbor antimicrobial resistance genes that may be transferred to other susceptible intestinal strains.
KEYWORDS: Bacteroidesspp.;Parabacteroides distasonis;b-lactamase activity; Antimicrobial resistance; Resistance genes.
Nakano V, Nascimento e Silva A, Merino VRC, Wexler HM, Avila-Campos MJ. Antimicrobial resistance and prevalence of resistance genes in intestinal Bacteroidalesstrains. Clinics. 2011;66(4):543-547.
Received for publication onSeptember 11, 2010;First review completed onOctober 18, 2010;Accepted for publication onDecember 17, 2010
E-mail: vivinkn@usp.br
Tel.: 55 11 30917344
INTRODUCTION
Bacteroides andParabacteroidesspecies are components of the colon resident microbiota, and both genera belong to the order Bacteroidales.1 Species of these genera are often
associated with opportunistic mixed infections, such as intra-abdominal, obstetric-gynecologic and diabetic foot infections. In addition, these microorganisms are able to develop resistance to several antimicrobial drugs.2
Although antibiotics with good activity against these bacteria are currently available, high frequencies of resis-tance to some antimicrobials have been reported in several countries.2,3
BacteroidesandParabacteroidesspecies produce endogenous
b-lactamases, the most important mechanism of resistance to
b-lactam antibiotics.Bacteroides fragilisis the most frequently isolated bacteria from infectious diseases, and it exhibits high
levels of resistance to b-lactam drugs4compared with other
Bacteroidales because of the production of cephalosporinases and penicillinases encoded by thecepAgene.5
Bacterial resistance to imipenem, ertapenem and merope-nem arises because of the production of metallo-b-lactamase (class B) encoded by thecfiAgene, but this resistance is rarely observed in Bacteroidesand Parabacteroidesspecies.6Strains harboring ‘‘silent’’cepAorcfiAgenes appear to be resistant to penicillin, cephalosporin or carbapenem. Conversely, some
B. fragilis strains harboring either cepA or cfiA genes are susceptible to b-lactams, but after antibiotic pressure, they become resistant because of an insertion sequence (IS) in the upstream region of these genes.7
Clindamycin resistance rates have been shown to vary from 10% to 42% in intestinal Bacteroidales strains world-wide.3,8 Clindamycin resistance is encoded by the ermF
gene, which confers resistance to macrolides, lincosamides and streptogramin B via a 23S rRNA mechanism, which produces the methylases ErmF, EmFS, ErmG and ErmB.9
Metronidazole resistance in anaerobic bacteria appears to be associated with thenim AtoGgenes that are transcribed by promoters located in different IS-producing nitroimida-zole reductases, which transform 4- or 5-nitroimidanitroimida-zole Copyrightß2011CLINICS– This is an Open Access article distributed under
genes to 4- or 5-aminoimidazoles.10 Nitroimidazole resis-tance inBacteroidesspp. andParabacteroides distasonisis not commonly observed. Although non-nim genes associated with imidazole resistance have been reported, this resis-tance might be due to an extensive use of metronidazole. However, the exact mechanism of this resistance remains undefined.11
The aim of this study was to determine the antimicrobial resistance profile and the prevalence of resistance genes in
Bacteroidesspp. andParabacteroides distasonisstrains isolated from children’s intestinal microbiota.
METHODS AND MATERIALS
Bacteria
A total of 114 intestinalBacteroidalessamples (66Bacteroides fragilis; 14B. vulgatus; 7B. uniformis;7B. ovatus;2B. eggerthii; 2
B. thetaiotaomicronand 16Parabacteroides distasonis) isolated from 39 fecal samples from children were evaluated. Children from 2 children’s hospitals and 2 day care centers (Sa˜o Paulo, SP, Brazil) were selected for this study, with ages ranging from 2 months to 8 years old. None of the subjects received antibiotic therapy prior to sample collection. Fecal samples were collected from April to December 2000. Stools were plated onto Bacteroides fragilis-bile-esculin agar and identified using an established methodology12. The strains were stored at -80
˚
C in 10% skim milk. This study was approved by the Ethics Commission of the Instituto de Cieˆncias Biome´dicas, USP (158/CEP).Susceptibility Testing
Antimicrobial susceptibility tests were performed using an agar dilution method in Wilkins & Chalgren agar in accordance with the recommendations of the Clinical and Laboratory Standards Institute (CLSI).13 The antibiotics used were as follows: amoxicillin, ampicillin, cephalexin, clindamycin and tetracycline (Luper Ind. Farm. Ltd., SP, Brazil), cefoxitin and imipenem (Merck Sharp & Dohme, SP), amoxicillin/clavulanic acid (Smithkline Beechman Brazil Ltd., SP), metronidazole (Aventis Farm. Ltd., SP) and penicillin (Prodoti Lab. Farm. Ltd., SP). Briefly, media containing twofold serial dilutions of antimicrobial agents ranging from 0.25 to 512 mg/ml were inoculated with 1.56105cfu delivered by a Steers replicator. Media without
antibiotics were used as controls. Plates were incubated in anaerobic conditions (90% N2/10% CO2) at 37
˚
C for 48 h.The Minimal Inhibitory Concentration (MIC) was defined as the lowest concentration of each antimicrobial agent able to inhibit visible bacterial growth. All tests were performed in duplicate. The B. fragilis ATCC 43858 was included as a control in all assays to assess the reliability of the methods.
Determination ofb-lactamase activity
Hydrolysis of the chromogenic cephalosporin (Nitrocefin, Oxoid Ltd., Sa˜o Paulo, SP, Brazil) was used to observe enzyme production. b-lactamase activity was expressed semi-quantitatively; negativeb-lactamase activity was indi-cated by a yellow color and positive by a red color. The penicillin-resistant andb-lactamase-positive strainB. fragilis
ATCC 43858 was used as a control.
Detection of resistance genes by a PCR assay Bacterial genomic DNA was obtained using an Easy-DNA kit (Invitrogen do Brasil Ltd., Sa˜o Paulo, SP, Brazil)
according to the manufacturer’s instructions. PCR assays were used to detect the presence of resistance genes (cfiA,
cepA, ermF, tetQandnim) and the insertion sequences (IS942
andIS1186) associated withcfiA gene expression.5DNA amplifications were performed in volumes of 25mL contain-ing 1 X PCR buffer (Invitrogen), 2.5 mM MgCl2, 0.2 mM
dNTP mix (Invitrogen), 0.4mM of each primer (Invitrogen), 0.5 U of Platinum Taq DNA polymerase (Invitrogen and 10 ng of DNA. Amplifications were performed in a thermal cycler (PerkinElmer Amp PCR System 9700). Table 1 shows the PCR conditions, including the genes, primer sequences and cycles. Amplification products were analyzed by electrophoresis in a 1% agarose gel (in 1X TBE buffer), stained with ethidium bromide and photographed under UV light.
Statistical Analysis
All statistical analyses were performed using GraphPad InStat statistical analysis software (version 3.05, GraphPad Software) with a one-way ANOVA. A difference ofp#0.05
was considered statistically significant.
RESULTS
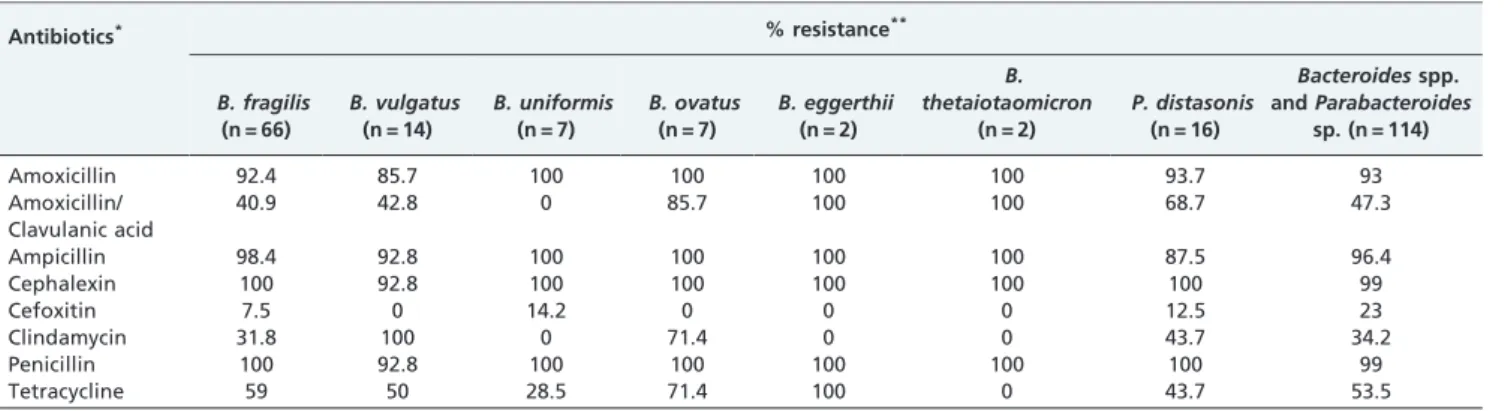
All tested strains were susceptible to imipenem and metronidazole. Cefoxitin was active against 77% of the tested bacteria. In addition, clindamycin was active against 65.8% of the tested bacteria, combined amoxicillin/clavu-lanic acid against 52.7%, and tetracycline against 46.4%. All intestinalBacteriodalesspecies exhibited antimicrobial resis-tance ranging from 23% to 99% (Table 2). Most strains (92%) were able to produce b-lactamases (Table 3). All strains harbored at least one of the resistance genes evaluated. For
Table 1- Resistance genes, oligonucleotide sequences and PCR conditions used to detect target genes
Resistanc Genes
Oligonucleotide Sequence 59R39
Amplification
Cycles Reference
cepA TTT CTG CTA TGT CCT GCC C 35 cycles: 5 ATC TTT CAC GAA GAC GGC 94˚C660 sec
52˚C660 sec 72˚C660 sec
cfiA ATG GTA CCT TCC AAC GGG 35 cycles: 5 CAC GAT ATT GTC GGT CGC 94˚C660 sec
56˚C660 sec
72˚C660 sec
IS1186 TGA CCT ACA ACA TCT TCC G 35 cycles: 5 GGT TGT TGA TAA CAA TCA
TCC C
94˚C660 sec
50˚C660 sec
72˚C62 min
IS942 TCC TCA ATA CAT GAG CCG C 35 cycles: 5 GGT TGT TGA TAA CAA TCA
TCC C
94˚C660 sec
50˚C660 sec
72˚C62 min
tetQ ACT TCC GTA ACC GAG AAT CTG CTG TAC CGG ATA GAC TTT GGC
TTT TGC
40 cycles: 94˚C660 sec
50˚C660 sec
72˚C640 sec
30
ermF CGG GTC AGC ACT TTA CTA TTG
GGA CCT ACC TCA TAG ACA AG
35 cycles: 94˚C630 sec
50˚C630 sec 72˚C62 min
23
nim ATG TTC AGA GAA ATG GGG CGT AAG CG GCT TCC TTG CCT GTC ATG
TGC TC
34 cycles: 94˚C660 sec
55˚C660 sec
72˚C630 sec
example, 71 strains (62.3%) harbored thecfiAgene and were susceptible to imipenem. Moreover, mobile elements were observed in 2 B. fragilis (IS1186 and IS942) strains, 1 B. vulgatus(IS1186) strain and 1P. distasonis(IS942) strain, but none carried thecfiAgene. ThecepAgene was present in 87 (76.3%) of the tested Bacteroidales species, and high resistance values to some antimicrobials, including cepha-lexin and penicillin (99%), ampicillin (96.4%), and amox-icillin (93%), were observed, suggesting the possibility of an association between the presence of these genes and the resistance to cephalosporin and penicillin. Out of 39 clindamycin-resistant strains, 31 (79.5%) harbored theermF
gene. While 91 (79.8%) of the tested strains harbored the
tetQ gene, only 61 (67%) were resistant to tetracycline. Bacterial strains were susceptible to metronidazole. However, 9 strains (5B. fragilis, 1B. vulgatus, 2B. uniformis
and 1P. distasonis) harbored thenimgene. The presence of resistance genes in all testedBacteroides andParabacteroides
strains was statistically significant (p,0.001), and p,0.01 was observed in B. fragilis strains. Table 3 shows the distribution of the resistance genes inBacteroidesspp. andP. distasonis.
DISCUSSION
Bacteroidales species are important anaerobe components of the resident intestinal microbiota, and they are potential endogenous pathogens.Bacteroidesspecies andP. distasonis
have been shown to induce different infections in humans.2
These intestinal anaerobes are resistant to several penicillins and cephalosporins,14 but the exact mechanism of this
resistance is unknown.
In this study, a high rate ofb-lactamase-producing strains (92%) was observed in accordance with previous studies.3,15
In addition, some resistant strains can produceb-lactamases that are encoded by plasmid-borne or chromosomal cepA
genes, and these enzymes are responsible for the increase in antibiotic resistance.
Most anaerobic bacteria are susceptible to imipenem,4,17 although high rates of resistance to this drug have been reported.2,14 Moreover, in Bacteroidales the cfiA gene has been detected at a low rate.16,18,19In this study, 71 (62.3%) of the tested strains harbored thecfiAgene, but no imipenem-resistant strains were observed. Conversely, high detection rates of the cfiA gene suggest that these strains act as reservoirs for antibiotic resistance genes, which is in accordance with the results of Garcia et al.20 The role of thecfiAgene in these intestinal strains remains unclear, and further studies are necessary to understand its presence. Moreover,cfiA-positive Bacteroidales strains did not harbor either IS942 or IS1186 elements, which has also been demonstrated by Soki et al.18and Walsh et al.21In addition,
strains susceptible to imipenem did not harbor the IS promoter.
The production of cephalosporinases and penicillinases
encoded by the cepA gene is commonly observed in
Bacteroidesspp. andParabacteroides distasonis.5In this study, 87 (82.8%) of 105 b-lactamase-producing strains harbored Table 2 -Resistance profiles of intestinalBacteroidalesspecies to 8 antibiotics.
Antibiotics* % resistance**
B. fragilis (n = 66)
B. vulgatus (n = 14)
B. uniformis (n = 7)
B. ovatus (n = 7)
B. eggerthii (n = 2)
B. thetaiotaomicron
(n = 2)
P. distasonis (n = 16)
Bacteroidesspp. andParabacteroides
sp. (n = 114)
Amoxicillin 92.4 85.7 100 100 100 100 93.7 93
Amoxicillin/ Clavulanic acid
40.9 42.8 0 85.7 100 100 68.7 47.3
Ampicillin 98.4 92.8 100 100 100 100 87.5 96.4
Cephalexin 100 92.8 100 100 100 100 100 99
Cefoxitin 7.5 0 14.2 0 0 0 12.5 23
Clindamycin 31.8 100 0 71.4 0 0 43.7 34.2
Penicillin 100 92.8 100 100 100 100 100 99
Tetracycline 59 50 28.5 71.4 100 0 43.7 53.5
*Breakpoints used in accordance with CLSI (2007): Amoxicillin (8mg/mL); Amoxicillin/clavulanic acid (8mg/mL); Ampicillin (1mg/mL); Cephalexin (8mg/mL); Cefoxitin (32mg/mL); Clindamycin (4mg/mL); Imipenem (8mg/mL); Metronidazole (16mg/mL); Penicillin (1mg/mL) and Tetracycline (8mg/mL).
**All strains were susceptible to imipenem and metronidazole.
***B. fragilisATCC 43858 was resistant to amoxicillin, ampicillin, cephalexin, clindamycin and penicillin.
Table 3 -Distribution of resistance genes andb-lactamase production in intestinalBacteroidesspp. andP. distasonis.
Species (n) Genes b-lactamase production
cfiA cepA ermF tetQ nim
n˚(%) n˚(%) n˚(%) n˚(%) n˚(%) n˚(%)
B. fragilis(66) 51 (77.2) 53 (80.3) 16 (24.2) 54 (81.8) 5 (7.5) 60 (90.9)
B. vulgatus(14) 5 (35.7) 11 (78.5) 5 (35.7) 7 (50) 1 (7.14) 13 (92.8)
B. uniformis(7) 6 (85.7) 7 (100) 0 (0) 5 (71.4) 2 (28.5) 7 (100)
B. ovatus(7) 1 (14.2) 1 (14.2) 1 (14.2) 6 (85.7) 0 (0) 7 (100)
B. eggerthii(2) 0 (0) 2 (100) 1 (50) 2 (100) 0 (0) 2 (100)
B. thetaiotaomicron(2) 2 (100) 2 (100) 2 (100) 2 (100) 0 (0) 1 (50)
P. distasonis(16) 6 (37.5) 11 (68.7) 6 (37.5) 15 (93.7) 1 (6.25) 15 (93.7)
the cepA gene, suggesting that some strains were able to produce this enzyme using mechanisms other than the cepA gene.
Most b-lactamase-producing strains were susceptible to cefoxitin, with a resistance rate of only 23% (Table 2). These data are supported by previously published studies.3,8 Moreover, the combination of amoxicillin and clavulanic acid did not show good activity against 47.3% of the tested strains, in accordance with Wybo et al.3and Roberts et al.17 Clindamycin is a semi-synthetic drug used extensively in the treatment of anaerobic infections.22 However, bacterial resistance to this drug has significantly increased over the last two decades. In this study, 34.2% of strains were observed to be clindamycin resistant, in accordance with Betriu et al.14IntestinalBacteroidalesstrain resistance rates to clindamycin have been shown to vary between countries from 39% to 41%.3,4,16
The ermB, ermF, ermG, and ermS genes are the most common determinants of genetic resistance in intestinal
Bacteroidales strains.23 Of the 39 (34.2%)
clindamycin-resistant strains in this study, only 10 harbored the ermF
gene. Bacterial resistance to clindamycin can arise because of the presence of the ermB or ermG genes24 or by other mechanisms, such as efflux pumps.25 In addition,
clinda-mycin resistance amongBacteroidales species has increased in several countries.26,27 This alarming resistance to clin-damycin amongBacteroidesspp. andP. distasonismakes its use unacceptable for the empiric therapy of severe anaerobic infections.
Tetracycline is one of the most widely used antibiotics worldwide, but its use has decreased because of the high resistance rates observed in various microorganisms, including Bacteroides spp. and Parabacteroides distasonis.16 Efflux pumps, ribosome protection and tetracycline mod-ification are the main mechanisms of bacterial resistance to tetracycline. However, ribosome protection appears to be the most widespread in nature.28ThetetQandtetMgenes encoding the ribosome-protecting proteins are often asso-ciated with conjugative transposons.29In this study, 53.5% of the tested strains were resistant to tetracycline, and among the 91 (79.8%) of 114 total strains harboring thetetQ
gene, 61 (67%) showed resistance to tetracycline. This result suggests that these bacteria may become resistant either by activating other genes, such astetM,tetK,tetLandtetO, or by another mechanism of resistance that remains to be clarified. All tested strains were susceptible to metronidazole in accordance with Odou et al.4However, other studies have noted an increase in the rate of resistance to metronida-zole.3,11In this study, only 9 (7.8%) strains harbored thenim
gene. Metronidazole resistance associated withnimhas been described in Bacteroides spp. and Parabacteroides distasonis
strains from different geographic regions.11 However,
resistance to metronidazole does not depend on the presence ofnimgenes, and the true role of these genes is not yet clear. In addition, nim-negative strains expressing high levels of resistance to metronidazole have been sporadically isolated, suggesting an additional mechanism of resistance11 and also justifying additional studies concerning the susceptibility profile and detection of nim
genes.
Few studies have addressed antimicrobial susceptibility profiles and the detection of resistance genes in intestinal anaerobic resident microbiota in Brazil, especially with a focus on children. Careful monitoring of antimicrobial
resistance and detection of these genes might be of interest, verifying the presence and spread of intestinalBacteriodales
strains with resistance markers to different antimicrobials in different countries.
ACKNOWLEDGEMENTS
The authors thank Mrs. Zulmira Alves de Souza for her technical support. This study was supported by Fundac¸a˜o de Amparo a Pesquisa do Estado de Sa˜o Paulo (FAPESP Grant 08/57330-4 and 09/03792-0).
REFERENCES
1. Sakamoto M, Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae com. nov. Int J Syst Evol Microb. 2006;56:1599-605, doi: 10.1099/ijs.0.64192-0.
2. Snydman DR, Jacobus NV, McDermott LA, Ruthazer R, Golan Y, Goldstein EJC, et al. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends in the United States from 1997 to 2004. Antimicrob Agents Chemother. 2007;51:1649-55, doi: 10. 1128/AAC.01435-06.
3. Wybo I, Pierard D, Verschraegen I, Reynders M, Vandoorslaer K, Clayes G, et al. Third Belgian multicentre survey of antibiotic susceptibility of anaerobic bacteria. J Antimicrob Chemother. 2007;59:132-9, doi: 10.1093/ jac/dkl458.
4. Odou MF, Muller C, Calvet L, Dubreuil L. In vitro activity against anaerobes of retapamulin a new topical antibiotic for treatment of skin infections. J Antimicrob Chemother. 2007;59:646-51, doi: 10.1093/jac/dkm019. 5. Gutacker M, Valsangiacomo C, Piffaretti JC. Identification of two genetic
groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology. 2000;146:1241-4.
6. Papaparaskevas J, Pantazatou A, Katsandri A, Legakis NJ, Avlamis A. Multicentre survey of the in-vitro activity of seven antimicrobial agents, including ertapenem, against recently isolated Gram-negative anaerobic bacteria in Greece. Clin Microbio Infect. 2005;11:820-4, doi: 10.1111/j. 1469-0691.2005.01233.x.
7. Podglajen I, Breuil J, Rohaut A, Monsempes C, Collatz E. Multiple mobile promoter regions for the rare carbapenem resistance gene of Bacteroides fragilis. J Bacteriol. 2001;183:3531-5, doi: 10.1128/JB.183.11. 3531-3535.2001.
8. Ulger NT, Gu¨lliioglu BM, C¸ akici O, Akin L, Dermirkalem P, C¸ elenk T, et al. Do antimicrobial susceptibility patterns of colonic isolates of Bacteroides species change after antibiotic prophylaxis with cefoxitin during elective abdominal surgey? World J Surg. 2005;29:1311-5, doi: 10. 1007/s00268-005-7961-3.
9. Gupta A, Vlamakis H, Shoemaker N, Salyers AA. A new Bacteroides conjugative transposon that carries an ermB gene. Appl Environ Microbiol. 2003;69:6455-63, doi: 10.1128/AEM.69.11.6455-6463.2003. 10. Haggoud A, Reysset G, Azeddoug H, Sebald M. Nucleotide sequence
analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob Agents Chemother. 1994;38:1047-51.
11. Lo¨fmark S, Edlund C, Nord CE. Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin Infect Dis. 2010;50:16-23, doi: 10.1086/647939.
12. Jousimies-Somer H, Summanen P, Citron DM, Baron EJ, Wexler HM, Finegold SM. Wadsworth-KTL Anaerobic Bacteriology Manual. 6 th ed. Belmont, CA: Star Publishing; 2002.
13. Clinical and Laboratory Standards Institute. Methods for Antimicrobial Susceptibility Testing of Anaerobic Bacteria. 7 th ed., Approved Standard M11-A7. CLSI, Wayne, PA, US, 2007.
14. Betriu C, Culebras E, Go´mez M, Lo´pez F, Rodrı´guez-Avial I, Picazo JJ. Resistance trends of the Bacteroides fragilis group over a 10-year period, 1997 to 2006, in Madrid, Spain. Antimicrob Agents Chemother. 2008;52: 2686-90, doi: 10.1128/AAC.00081-08.
15. Ca´ceres M, Zhang G, Weintraub A, Nord CE. Prevalence and antimicrobial susceptibility of enterotoxigenic Bacteroides fragilis in children with diarrhea in Nicaragua. Anaerobe. 2000;6:143-8, doi: 10. 1006/anae.2000.0341.
16. Paula GR, Falca˜o LS, Antunes ENF, Avelar KES, Reis FNA, Maluhy MA, et al. Determinants of resistance inBacteroides fragilisstrains according to recent Brazilian profiles of antimicrobial susceptibility. Int J Antimcrob Agents. 2004;24:53-8, doi: 10.1016/j.ijantimicag.2003.11.011.
18. So´ki J, Urban E, Fodor SE, Nagy E. Prevalence of the carbapenemase gene (cfiA) among clinical and normal flora isolates of Bacteroides species in Hungary. J Med Microbiol. 2000;49:427-30.
19. Ang L, Brenwald NP, Walker RM, Andrews J, Fraise A. Carbapenem resistance inBacteroides fragilis.J Antimicrob Chemother. 2007;59:1042-4, doi: 10.1093/jac/dkm062.
20. Garcı´a N, Gutie´rrez G, Lorenzo M, Garcı´a JE, Pı´riz S, Quesada A. Genetic determinants for cfxA expression in Bacteroides strains isolated from human infections. J Antimicrob Chemother. 2008;62:942-7, doi: 10.1093/jac/dkn347. 21. Walsh TR, Onken A, Haldorsen B, Toleman MA, Sundsfjord A. Characterization of a carbapenemase-producing clinical isolates of Bacteroides fragilis in Scandinavia: genetic analysis of a unique inser-tion sequence. Scand J Infect Dis. 2005;37:676-9, doi: 10.1080/ 00365540510034482.
22. Heddberg M, Nord CE. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe. Clin Microbiol Infect. 2003;9:475-88, doi: 10.1046/j.1469-0691.2003.00674.x.
23. Chung WO, Werckenthin C, Schwarz S, Roberts MC. Host range of the ermF rRNA methylase gene in bacteria of human and animal origin. J Antimicrob Chemother. 1999;43:5-14, doi: 10.1093/jac/43.1.5. 24. Shoemaker NB, Vlamakis H, Hayes K, Salyers AA. Evidence for
extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl Environ Microbiol. 2001;67:561-8.
25. Pumbwe L, Wareham DW, Aduse-Opoku J, Brazier JS, Wexler HM. Genetic analysis of mechanisms of multidrug resistance in a clinical isolate of Bacteroides fragilis. Clin Microbiol Infect. 2007;13:183-9, doi: 10. 1111/j.1469-0691.2006.01620.x.
26. Hedberg W, Shahin M, Rotimi VO. Antimicrobial susceptibilities of Bacteroides fragilis group isolates in Europe. Clin Microbiol Infect, 2003; 9:475-88, doi: 10.1046/j.1469-0691.2003.00674.x.
27. Jamal W, Shahin M, Rotimi VO. Surveillance and trends of antimicrobial resistance among clinical isolates of anaerobes in Kuwait hospitals from 2002 to 2007. Anaerobe. 2010;16:1-5, doi: 10.1016/j.anaerobe.2009.04.004. 28. Nickolich MP, Shoemaker NB, Salyers AA. A Bacteroides tetracycline resistance gene represents a new class of ribosome protection tetracycline resistance. Ant Agents Chemother. 1992;36:1005-12.
29. Leng Z, Riley DE, Berger RE, Krieger JN, Roberts MC. Distribution and mobility of the tetracycline resistance determinant tetQ. J Antimicrob Chemother. 1997;40:551-9, doi: 10.1093/jac/40.4.551.
30. Manch-Citron JN, Lopez GH, Dey A, Rapley JW, MacNeill SR, Cobb CM. PCR monitoring for tetracycline resistance gene in subgingival plaque following site-specific periodontal therapy. J Clin Periodontol. 2000;27:437-46, doi: 10.1034/j.1600-051x.2000.027006437.x.