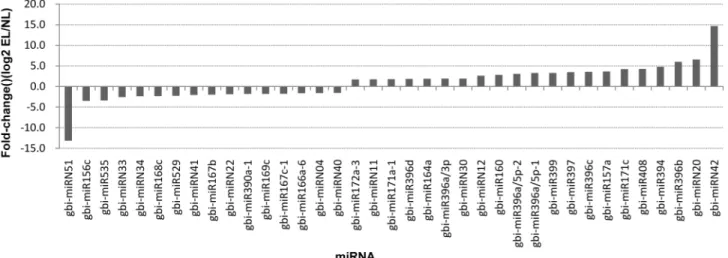
Identification and Characterization of MicroRNAs in Ginkgo biloba var. epiphylla Mak.
Texto
Imagem

Documentos relacionados
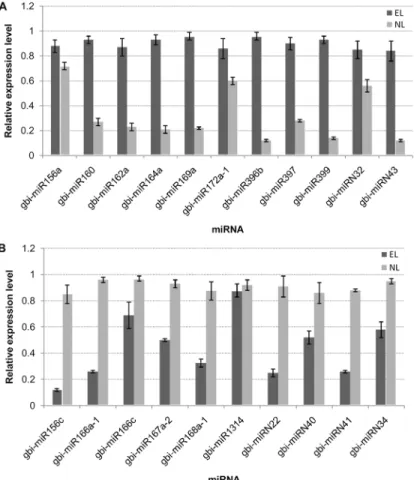
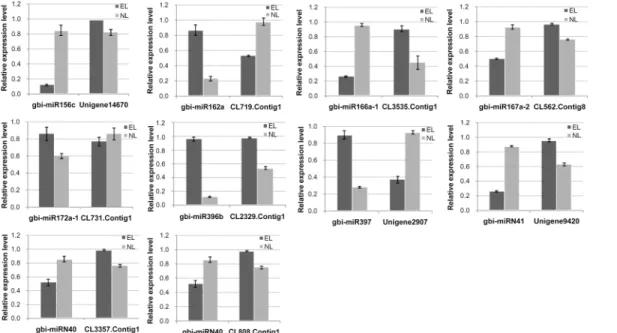
Statistical analysis - The expression levels of the miRNAs were expressed as the mean ± standard devia- tion. Means were compared via one-way ANOVA and the
Differential expression of 4 of 6 genes found to be differentially expressed by cDNA microarray analysis in the CSIII tumor versus normal tissue com- parison was validated
Expression profiles of GmIPT s and GmCKX s in the leaves, roots and root hairs of soybean plants at reproductive stage under normal and drought conditions.. Black bars; expression
The essential oils of leaves, twigs, branches, trunk bark and fruits of Croton palanostigma were analyzed by GC and GC-MS.. Statistical analysis showed that the leaves and
Em 2002, surge um outro sistema experimental de tradução, o TESSA [31] com o objetivo de auxiliar a comunicação entre uma pessoa surda e o balconista dos correios através da
RNA-Seq analysis and identification of differentially expressed genes (DEGs) - We performed RNA-Seq anal- yses to evaluate spatiotemporal gene expression profiles along
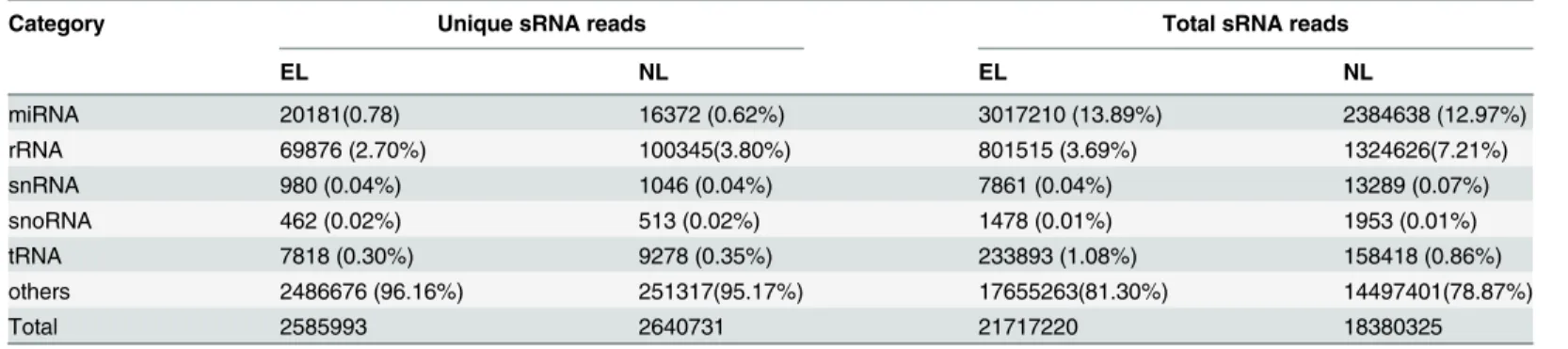
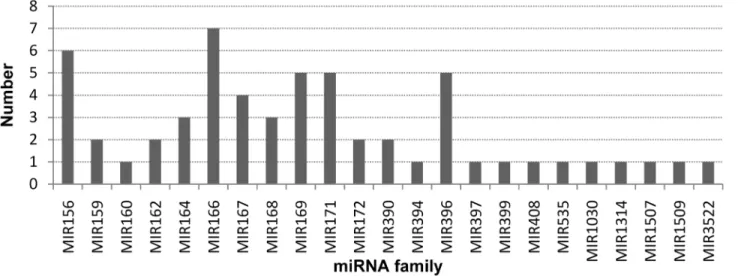
To iden- tify conserved miRNAs in wheat, we aligned the small RNA sequences against the known plant mature miRNAs registered in the miRBase (Release 21: June 2014), and
To profile differentially expressed lncRNAs in hepatoblastoma, we performed a genome-wide analysis of lncRNA and mRNA expression in hepatoblastoma and matched normal tissues