Degradation Pathway by
Rhodococcus
sp. Strain MB-P1
Fazlurrahman Khan, Deepika Pal, Surendra Vikram, Swaranjit Singh Cameotra*
Institute of Microbial Technology, Council of Scientific and Industrial Research (CSIR), Chandigarh, India
Abstract
2-chloro-4-nitroaniline (2-C-4-NA) is used as an intermediate in the manufacture of dyes, pharmaceuticals, corrosion inhibitor and also used in the synthesis of niclosamide, a molluscicide. It is marked as a black-listed substance due to its poor biodegradability. We report biodegradation of 2-C-4-NA and its pathway characterization byRhodococcussp. strain MB-P1 under aerobic conditions. The strain MB-P1 utilizes 2-C-4-NA as the sole carbon, nitrogen, and energy source. In the growth medium, the degradation of 2-C-4-NA occurs with the release of nitrite ions, chloride ions, and ammonia. During the resting cell studies, the 2-C-4-NA-induced cells of strain MB-P1 transformed 2-C-4-NA stoichiometrically to 4-amino-3-chlorophenol (4-A-3-CP), which subsequently gets transformed to 6-chlorohydroxyquinol (6-CHQ) metabolite. Enzyme assays by cell-free lysates prepared from 2-C-4-NA-induced MB-P1 cells, demonstrated that the first enzyme in the 2-C-4-NA degradation pathway is a flavin-dependent monooxygenase that catalyzes the stoichiometric removal of nitro group and production of 4-A-3-CP. Oxygen uptake studies on 4-A-3-CP and related anilines by 2-C-4-NA-induced MB-P1 cells demonstrated the involvement of aniline dioxygenase in the second step of 2-C-4-NA degradation. This is the first report showing 2-C-4-NA degradation and elucidation of corresponding metabolic pathway by an aerobic bacterium.
Citation:Khan F, Pal D, Vikram S, Cameotra SS (2013) Metabolism of 2-Chloro-4-Nitroaniline via Novel Aerobic Degradation Pathway byRhodococcussp. Strain MB-P1. PLoS ONE 8(4): e62178. doi:10.1371/journal.pone.0062178
Editor:Stephen J. Johnson, University of Kansas, United States of America
ReceivedJanuary 10, 2013;AcceptedMarch 18, 2013;PublishedApril 17, 2013
Copyright:ß2013 Khan, et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Funding:The authors have no support or funding to report.
Competing Interests:The authors have declared that no competing interests exist. * E-mail: ssc@imtech.res.in
Introduction
2-Chloro-4-nitroaniline (C6H5ClN2O2, 2-C-4-NA) is a
nitroaro-matic compound, used as an intermediate in the synthesis of dyes, pharmaceuticals, corrosion inhibitors and in the manufacture of niclosamide, a molluscicide [1–3]. 2-C-4-NA is also reported as a photolysis product of niclosamide [4]. Espinosa-Aquirre et al. [5] reported the metabolism of niclosamide, which is used as an anti-helminthic drug, results in the formation of 2-C-4-NA and 5-chlorosalicylic acid metabolites by hydrolytic cleavage of amide bond. As a result of its extensive production and application it may get released into the environments through various waste streams and is considered to be an increasing threat into the environments and various life forms [6]. Hence the fate of 2-C-4-NA in the environments is of great concern. 2-C-4-NA causes severe cellular damage as studied in rat [7]. It is identified as a bacterial mutagenic compound as tested inSalmonella typhimurium, however, the potency is low compared to niclosamide [5]. The environ-mental fate of 2-C-4-NA can be determined either by non-biological (volatization, hydrolysis, photolysis, thermal decompo-sition) or by biological means. Volatization of 2-C-4-NA from the soil surface and water surface are not considered to be an important fate process owing to its Henry’s Law constant of 9.561029 atm-cu m mole21 [8]. Similarly, 2-C-4-NA does not
contain any functional group by which it would hydrolyze in the environment [9]. However, 2-C-4-NA contain chromophores that can absorb light at wavelength of .290 nm and may be susceptible to photolysis [9]. Thus, for the remediation of the heavy contamination in the environment, the microbial
transfor-mation and degradation could be used as the most effective, eco-friendly and technically challenging approach for the decontam-ination of soil, sediment and water bodies etc. 2-C-4-NA is considered to be non-biodegradable in the aquatic environment as well as in the industrial sewage treatment plants [10]. Similarly, Canton et al. [11] also classified 2-C-4-NA as a blacklist substance due to its poor biodegradability. However, based on the previous reports on the biodegradation of structural analogues of 2-C-4-NA such as 4-nitroaniline, 2-, 3-, 4-chloroaniline and 3, 4-dichloroani-line, possibilities for the biodegradation of 2-C-4-NA by microor-ganisms could also be presumed [12–18]. In this communication, we report metabolic characterization of 2-C-4-NA byRhodococcus
Materials and Methods
Chemicals, strain, and growth medium
Analytical grade of 2-chloro-4-nitroaniline (2-C-4-NA) and standard 4-amino-3-chlorophenol (4-A-3-CP) were purchased from Sigma-Aldrich (St, Louis, MO, USA).Rhodococcus sp. strain MB-P1 was isolated from the contaminated soil sample and characterized for atrazine degradation [19]. Minimal salt medium (MSM) used in the present study was prepared as described earlier [19] with slight modification i. e. absence of nitrogen source [(NH4)2SO4]. Stock solution (10 mM) of 2-C-4-NA prepared in
HPLC grade methanol was added to an empty Erlenmeyer flask to obtain the working concentrations. Further, the residual methanol in the flask was evaporated under a stream of air to leave the dry crystal of 2-C-4-NA in the bottom of the flask. Appropriate volume of MSM was added to the flask to attain desired working culture. Nutrient agar at one-quarter strength (1/4-NA) and nutrient broth (1/4-NB) were used as a rich media for bacterial growth and culture maintenance.
Metabolic activity of strain MB-P1 on 2-C-4-NA
Metabolic activity of strain MB-P1 on 2-C-4-NA was deter-mined by growth studies carried out in carbon-free MSM supplemented with varying concentrations of 2-C-4-NA ranging from 50 to 500mM. The positive metabolic activity was determined by time dependent bacterial growth measure in terms of increase in optical density of the culture medium monitored at 600 nm using Lambda EZ 201 UV-visible spectrophotometer (Perkin-Elmer Inc, USA). Bacterial growth was also monitored by measuring the total protein of the cultures grown on 2-C-4-NA with Pierce BCA protein assay kit (Thermo Scientific, USA). Release of nitrite ions (NO22), chloride ions (Cl2), and ammonia
(NH3) in the growth medium and gradual decrease in
concentra-tion of 2-C-4-NA were monitored as the alternative methods for determination of metabolic activity of strain MB-P1 on 2-C-4-NA. Subsequent characterization was carried out to determine the kinetics of 2-C-4-NA degradation by strain MB-P1. Appropriate biotic and abiotic controls were included wherever necessary. Procedures used for growth studies and resting cell studies are as follows:
Growth studies
Growth studies were performed in 50 ml of carbon-free MSM supplemented with 2-C-4-NA (200mM) by inoculating 1% (v/v) of overnight culture of MB-P1 cells grown in 1/4-NB. Cultures were incubated on a rotary shaker at 200 rpm at 30uC. Samples were withdrawn at every 8 h to monitor bacterial cell growth, release of NO22, Cl2, NH3,and degradation of the growth substrate. Cell
growth was monitored spectrophotometrically as described above. Bacterial growth was also monitored by the indirect approach of measuring total protein concentration in the culture. Culture fluid samples (2.0 ml) were centrifuged at 8,0006g for 10 min to obtain cell-free culture supernatants that were subsequently analyzed for NO22, Cl2, and NH3release, depletion of growth substrate and
identification of degradation intermediates using methods de-scribed later. Non-inoculated flask and flask inoculated with heat killed cells of strain MB-P1 were used as abiotic control and negative control respectively.
Resting cell studies
Resting cell studies were performed with minor modification of the method described earlier [20]. Briefly, a seed culture (6%, v/v) of strain MB-P1 grown in NB inoculated into 1.6 L of 1/4-NB supplemented with 200mM of 2-C-4-NA and incubated at
30uC with aeration for 24 h (OD600, 1.3–1.4). The induced cells
were harvested by centrifugation at 8,0006g at room temperature for 10 minutes, washed twice with phosphate buffer (20 mM, pH 7.2) and suspended in 100 ml of carbon-free MSM. This suspension was divided into four aliquots of 25 ml each; one aliquot was heat killed by incubating on boiling water for 30 minutes which was later used as negative control. The other two aliquots were supplemented with 200mM of 2-C-4-NA and 4-A-3-CP separately. Each flask was then incubated at 30uC with shaking at 200 rpm. Similarly, another control for the above experiment was also made by suspending un-induced cells of strain MB-P1 in MSM supplemented with 200mM of 2-C-4-NA and 4-A-3-CP separately. Samples (2.0 ml) were withdrawn from both control and experimental flasks at regular intervals of 2 h and subjected to NO22, Cl2, and NH3 release analysis, followed by
high-performance liquid chromatography (HPLC) analysis and also gas chromatography-mass spectroscopy (GC-MS) analyses (meth-ods described later).
Ring cleavage inhibition studies
There is a well known study to check the inhibition of ring cleavage catalyzed by ferrous ions dependent ring cleavage dioxygenase by using an iron chelator i. e. 2, 2-dipyridyl [21]. The ring cleavage experiment was performed in the same way as the resting cell study. The harvested 2-C-4-NA-induced resting cells were suspended in 25 ml carbon-free MSM supplemented with 200mM of 2-C-4-NA and 1.0 mM 2, 2-dipyridyl. Similarly, another flask was also taken as a control containing 25 ml cell suspensions of 2-C-4-NA-induced resting cells in MSM supple-mented with 200mM of 2-C-4-NA only. Each flask was then incubated at 30uC with shaking at 200 rpm. Culture supernatant (2.0 ml) was collected at different time intervals and analyzed by HPLC.
Enzyme assays with cell-free lysates
2-C-4-NA-induced cells of strain MB-P1 were harvested by centrifugation and were washed twice with phosphate buffer (20 mM, pH 7.2) and finally re-suspended in lysis buffer (50 mM phosphate buffer, pH 7.2). Cell lysis was carried out by two passages through a French pressure cell (20,000 lb/in2). The lysed cell suspensions were centrifuged at 12,000 rpm for 30 min at 4uC and supernatant was carefully separated to obtain cell-free enzyme extract, which was subsequently used for the enzyme assays. Quantitation of protein content within cell-free extracts were performed routinely with Pierce BCA protein assay kit (Thermo Scientific, USA). The cell-free extract was tested for activity of monooxygenase since this enzyme is known to be involved in the first step of 2-C-4-NA degradation as indicated by the identifica-tion of the pathway metabolites. The monooxygenase enzyme assay was carried out as described below:
Enzyme assays for monooxygenase
Flavin-dependent monooxygenase enzyme activity was assayed to characterize the putative first step of 2-C-4-NA degradation by strain MB-P1. Cell-free protein extracts (5 mg ml21
of protein) prepared from 2-C-4-NA-induced cells were added to phosphate buffer (20 mM, pH 7.0), 100mM NADPH, 200mM FMN in a total reaction volume of 10 ml. Reactions were initiated by the addition of 70mM 2-C-4-NA and incubated at 28uC. Samples were withdrawn at regular time intervals and monooxygenase enzyme activity was determined by measuring the time dependent disappearance of 2-C-4-NA and NO22 release. The 2-C-4-NA
depletion was measured by HPLC analysis. Release of NO22ions
Enzyme assay for aniline dioxygenase
The aniline dioxygenase enzyme activity by strain MB-P1 was measured with an oxygen electrode (YSI, Ohio, USA), according to the method described by Liu et al. [22]. MB-P1 cells grown in 500 ml 1/4-NB were harvested by centrifugation at 8,0006g at 4uC for 10 minutes. Pellets were washed twice with phosphate buffer (20 mM, pH 7.2) and suspended in 100 ml of MSM containing 2-C-4-NA (100mM). In order to obtain the un-induced cells, the similar cell pellets were suspended in 100 ml of 1/4-NB. Further, these cells were incubated at 30uC under shaking at 200 rpm to obtain 2-C-4-NA-induced or non-induced cells respectively. After 24 h of incubation, cells were harvested, cell pellets were washed twice with phosphate buffer (20 mM, pH 7.2) and re-suspended in the same buffer. This suspension was used for the assay of aniline dioxygenase by measuring oxygen uptake at 30uC.
Analytical methods
The release of nitrite ions (NO22) was monitored with a
colorimetric method using N-(1-naphthyl) ethylene-diamine-dihy-drochloride and sulfanilic acid reagent as described earlier [23]. Ammonia (NH3) concentrations were also monitored with a
colorimetric method using ‘Ammonia Estimation Kit’ (Sigma Aldrich, USA) according to the manufacturers’ recommendation. Similarly, the release of chloride ions (Cl2) was also monitored colorimetrically with colorimetric method that uses mercuric thiocyanate [24]. Standard plots generated with known concen-trations of NaCl, (NH4)2SO4and NaNO2were used to determine
the concentrations of Cl2, NH
4+, and NO22ions released into the
culture medium.
For the quantitative measurement of 2-C-4-NA degradation by strain MB-P1 and identification of metabolic products, 500ml of cell-free aqueous culture supernatants from growth and resting cell studies were filtered with 0.2mm filters (Millipore Inc. USA). The filtered culture supernatants were analyzed with minor modifica-tion of the quantitative HPLC method described earlier [25]. Briefly, the separation of 2-C-4-NA and their metabolic products was carried out on a C-18 reverse phase column at 30uC on UV-detector equipped Waters HPLC system (Waters, USA). The mobile phase used was a mixture of acetonitrile:water (30:70, v/v) under an isocratic condition with the flow rate of 1.0 ml min21
. The peaks of analytes were detected with UV detector over a wavelength scan of 220–290 nm. Alternatively for the analysis of metabolites by GC-MS, cell-free aqueous culture supernatant samples were mixed with equal volume of ethyl acetate and performed liquid-liquid extraction by layer separation sequentially at neutral and acidic pH. Extracted organic phase was pooled, dried under nitrogen flow using RotaVapor II (BUCHI, Switzer-land) and re-suspended in the appropriate volumes of ethyl acetate. The derivatization of metabolic products in the sample was performed by silylation as previously described [26]. The derivatized samples were analyzed by GC-MS using QP2010S (Shimadzu Scientific Instruments, USA) with temperature pro-gram and run parameter used for GC analyses as reported earlier [27]. Briefly, temperature program was: 1 min at 50uC, followed by ramping up to 150uC at a constant temperature increment rate of 20uC min21
, a slower ramping to 200uC at a rate of 5uC min21
, and then further ramped to 320uC at a rate of 20uC min21
, which was held for 5 min. Positive molecular ion mass spectra were scanned in mass/charge (m/z) range of 50–500.
Results
Growth study and degradation of 2-C-4-NA by strain MB-P1
Initial screening for the metabolic activity on 2-C-4-NA was carried out by using lab isolates (previously isolated from contaminated soil). A total 79 bacterial isolates were selected and screened for 2-C-4-NA degradation by inoculating in carbon-free MSM supplemented with 2-C-4-NA (100mM). Thereafter 2-C-4-NA degrading abilities were examined by analyzing the growth of culture and release of NO22, Cl2ions, and NH3in the
medium. Five isolates showed apparent increase in bacterial cell mass and also the release of NO22, Cl2ions, and NH3. Among
these isolates, strain MB-P1 was found to be an efficient degrader of 2-C-4-NA. The strain MB-P1 was incubated in carbon-free MSM supplemented with the different concentrations of 2-C-4-NA ranging from 50 to 500mM. Noticeably complete inhibition of growth was observed when 2-C-4-NA was used at concentrations higher than 300mM. However, 200mM served as optimal concentration for growth of strain MB-P1 (data not shown). The growth study of strain MB-P1 carried out in carbon-free MSM supplemented with 2-C-4-NA (200mM) showed an evident lag phase of 16 h (Figure 1). However, over time of subsequent log phase, the bacterial culture grew exponentially as observed with the increase of total protein concentration up to the value of 9.1mg ml21
(Figure 1). During the growth study, no metabolic products were detected from the sample analyzed by HPLC. However, NH3, Cl2ions, and NO22ions released in the culture media were
quantified colorimetrically. The released NH3 and NO22 ions
were quantified colorimetrically to be 30mM and 198.5mM respectively at the end of the log phase of the growth (Figure 1). The accumulation of lesser amount of NH3in the media clearly
suggested that NH3was used as a preferred nitrogen source for its
growth. On the other hand, Cl2ions continued to accumulate in culture medium and reached a maximum concentration of 198.8mM. The quantitative measurement of 2-C-4-NA by HPLC from growth medium showed its decrease from initial concentra-tion of 200mM to non-detectable amounts after,90 h of
incubation (Figure 1). The growth yield of strain MB-P1 on 2-C-4-NA was found to be 0.50 g of cells/g of 2-C-2-C-4-NA. Similarly, the rate of 2-C-4-NA degradation by strain MB-P1 during the growth studies was calculated to be 3.2mmol mg protein21minute21. The strain MB-P1 also utilized 4-A-3-CP as the sole carbon, nitrogen, and energy source with slight accumulation of NH3(28mM) in the
culture media (Data not shown). The growth yield of strain MB-P1 on A-3-CP was 0.62 g of cells/g of A-3-CP and the rate of 4-A-3-CP degradation by strain MB-P1 was 3.0mmol mg protein21
minute21
. The growth yield of strain MB-P1 on 4-A-3-CP as well as the rate of 4-A-3-CP degradation was found almost similar as determined for 2-C-4-NA.
Elucidation of catabolic pathway for 2-C-4-NA degradation by strain MB-P1
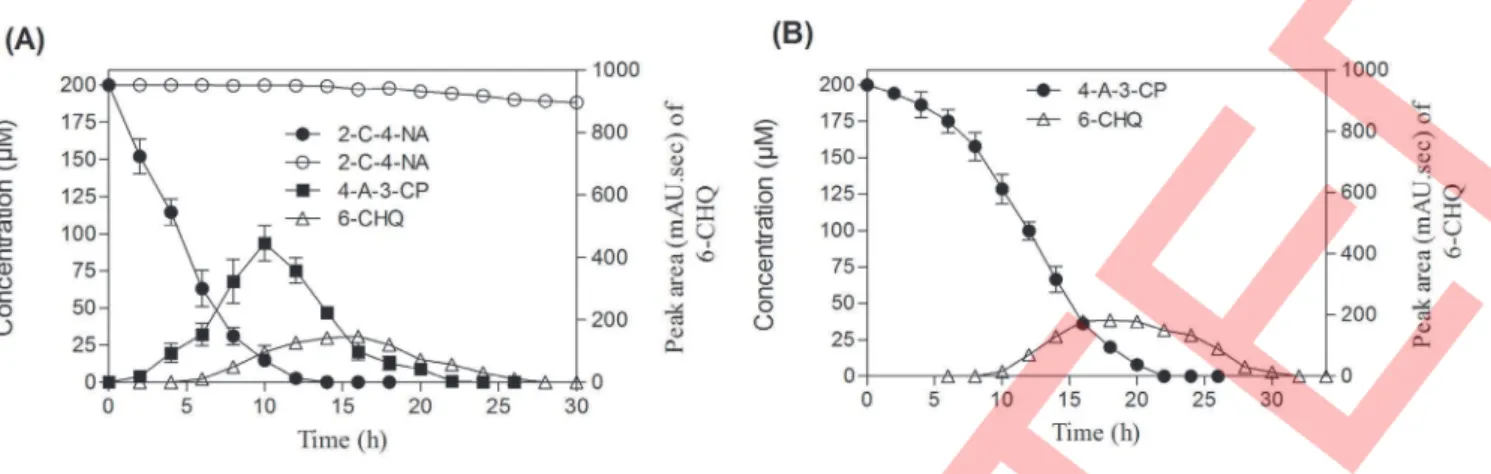
the disappearance of 4-A-3-CP in the first phase which underwent faster depletion over the time of incubation, suggesting that the enzyme responsible for the degradation of 4-A-3-CP is inducible in nature (Figure 2B). Noticeably, lesser amount of 6-CHQ metabolite was also identified, which disappeared over time of incubation (Figure 2). The above two metabolites identified in HPLC showed retention time (Rt) at 4.86 and 3.28 min respectively (Figure 3A). The Rt value corresponding to the 4.86 min matched with the authentic standard of 4-A-3-CP. However, the Rtvalue corresponding to the 3.28 min could not be characterized due to the non-availability of standard. Further, the confirmation and identification of unknown metabolites from the samples obtained from the resting cell study were subjected to mass fragmentation analysis by GC-MS. In GC-MS analysis two metabolic peaks appeared at Rt values of 7.62 and 9.52 min respectively (Figure 3B). The mass fragmentation pattern for metabolite with a Rtvalue of 7.62 min showed a conjugate ion at m/z value of 215 (representing silylated species: M+
), 200 (M-CH3), 143, 73 [Si (CH3)3] (Figure 3C). The mass spectrum of this
metabolite matched the known standard of 4-amino-3-chlorophe-nol. This metabolite would have probably resulted from direct denitrification of the aromatic nucleus without deamination. The second metabolite with a Rtvalue of 9.52 min showed a conjugate ion at m/z value of 378, 376 (M+), 366, 361 (M-CH
3), 288, 275,
273, 179, 73 [Si (CH3)3] (Figure 3C). The fragmentation pattern
and mass spectra of this metabolite were consistent and similar to that of chlorohydroxyquinol as described earlier [27]. 6-chlorohydroxyquinol is reported as the terminal intermediate in the degradation of 2, 4, 6-trichlorophenol and 2, 6-dichlorophenol degradation by Azotobacter sp. strain GP1, Streptomyces rochei 303,
Ralstonia eutrophaJMP 134 andCupriavidus necatorJMP134 [28–32]. The formation of 6-chlorohydroxyquinol metabolite would be as a result of dioxygenation with the deamination of amino group from the benzene ring. Based on the above, HPLC and GC-MS analysis, we conclude that 4-A-3-CP and 6-CHQ are the major identified metabolites during aerobic degradation of 2-C-4-NA by strain MB-P1.
Enzyme assays. To validate the formation of metabolites and illustrate the degradation pathway of 2-C-4-NA in strain MB-P1, the enzyme assays were carried out. The formation of 4-A-3-CP as the first degradation metabolite and the release of NO22
ions from 2-C-4-NA suggested oxygenase mediated enzymatic reaction in the degradation pathway. Cell lysates prepared from
2-C-4-NA grown cells of strain MB-P1 showed positive activity for flavin-dependent monooxygenase enzyme and catalyzed the elimination of NO22 ions and formation of 4-A-3-CP. The
specific activity for this enzymatic reaction was 1.360.25 nmol min21
mg of protein21
.
Oxygen uptake. There was rapid oxygen consumption by 2-C-4-NA, 4-nitroaniline and 4-chloroaniline by 2-C-4-NA-grown cells (Table 1). The oxygen uptake of the above results showed that the pathway enzymes involved in the degradation of 2-C-4-NA are induced. However, the lesser amount of oxygen consumption by 2-C-4-NA by 1/4-NB grown cells revealed the first enzyme is constitutive in nature. Whereas, there was a negligible amount of oxygen uptake by aniline and 4-A-3-CP in 1/4-NB grown cells indicating inducible nature of the aniline dioxygenase. The above results are in close agreement with the results as reported earlier [22,33,34]. The NH3 released were also quantitatively measured
and showed stoichiometry to the amount of substrates added (data not shown). Liu et al. [22] reported the formation of catechol product from aniline occurs via the action of aniline dioxygenase. Thus, based on the above results it is concluded that the second enzyme responsible for the degradation of 2-C-4-NA is a type of dioxygenase.
Ring cleavage inhibition studies
The HPLC analysis of the samples collected from the ring cleavage inhibition studies by 2, 2-dipyridyl on 2-C-4-NA showed the accumulation of 6-CHQ at the Rtvalue of 3. 28 minute with the liberation of only NO22 ions and NH3in the culture media
(Figure 4A). The culture media became deep red in color, which also indicates accumulation of hydroxyquinol as reported previ-ously [27,35]. Noticeably, a slight accumulation of 6-CHQ in the control (without added 2, 2-dipyridyl) was observed which got disappeared within a short time of incubation with the liberation of NO22, Cl2 ions, and NH3 (Figure 4B). The above results
clearly suggested that 6-CHQ is the terminal intermediate for the degradation of 2-C-4-NA by strain MB-P1. It is well known that the reaction mechanism of 2, 2-dipyridyl used as an inhibitor for the ring cleavage require ferrous ions for their enzymatic activities [21]. The formation of hydroxyquinol as a terminal ring cleavage substrate has been exclusively reported in the degradation pathway of 4-nitrophenol by gram-positive bacteria [35–37]. Similarly, the formation of 6-chlorohydroxyquinol has also been reported as a ring cleavage substrate in the degradation pathway of 4-chlorophenol [21]. Based on the identified metabolite from the resting cell study, oxygen uptake, enzyme assays and ring cleavage inhibition study, the pathway for the degradation of 2-C-4-NA has been proposed as shown in Figure 5.
Discussion
Biodegradation has been recognized as an economical and environment friendly approach for decontamination of sites polluted with toxic anthropogenic chemicals, released into environment as a result of industrial, agricultural, military and household activities [38,39]. Conversely, its application has remained elusive and inefficient in case of highly recalcitrant compounds including chloro and nitro containing anilines [14,40,41]. To overcome this limitation, it is essential to isolate and characterize microorganism(s) with potential to metabolize these compounds. To date no aerobic degradation and pathway characterization has been reported for the microbial degradation of 2-C-4-NA. However, based on the earlier reports on the degradation of structural analogues of 2-C-4-NA such as anilines substituted with chloro and nitro groups, it is presumed that the
Figure 1. Growth characterization ofRhodococcussp. strain MB-P1 on 2-C-4-NA as the sole carbon, nitrogen, and energy source. (
N
), 2-C-4-NA; (m), NO22; (D), NH42; (#), Cl2; (&), totalprotein. Values are presented as arithmetic means of data obtained from experiments carried out in triplicates; error bars represent standard deviation.
degradation of 2-C-4-NA could also be possible in aerobic condition. Although, the enrichment of cultures could be used as an efficient approach for isolating microorganism(s) with desired degradative capabilities; however, determining metabolic diversity of naturally occurring degradative isolates could also be another approach. Results presented in few recent studies indicate some
degradative strains to be metabolically versatile that they can degrade compounds analogous to their original enrichment substrate. The genus ofRhodococcusconstitutes a diverse group of bacteria which exhibit a metabolic versatile activity on chloro and nitro substituted aromatic compounds [42]. The catabolic versatility ofRhodococcus is due to the presence of large genome
Figure 2. Degradation kinetics of 2-C-4-NA and 4-A-3-CP during resting cell studies performed by 2-C-4-NA-induced cells of strain MB-P1.(A) 2-C-4-NA degradation kinetics. (
N
), 2-C-4-NA; (&), 4-A-3-CP; (D), 6-CHQ; (#), 2-C-4-NA in abiotic control. (B) 4-A-3-CP degradation kinetics. (N
), 4-A-3-CP; (D), 6-CHQ. Values are presented as arithmetic means of data obtained from experiments carried out in triplicates; error bars represent standard deviation.doi:10.1371/journal.pone.0062178.g002
Figure 3. Representative HPLC and GC-MS chromtograms along with mass spectra of metabolites identified during the degradation of 2-C-4-NA by resting cells ofRhodococcuss . (A) HPLC chromatograms of metabolites identified at different time intervals. Peak at Rtvalue of 8.5 min corresponds to 2-C-4-NA; Peak at Rtvalue of 4.86 min corresponds to 4-A-3-CP; Peak at Rtvalue of 3.28 min corresponds to 6-CHQ. (B) GC-MS chromatogram of the authentic standards and the identified metabolites. Peak at Rtvalue of 6.41 min corresponds to 2-C-4-NA; Peak at Rtvalue of 7.62 min corresponds to 4-A-3-CP; Peak at Rtvalue of 9.52 min corresponds to 6-CHQ. (C) Mass spectra of metabolites (Left side; 4-A-3-CP and right side; 6-CHQ) of 2-C-4-NA as analyzed by GC-MS.
doi:10.1371/journal.pone.0062178.g003
and plasmid which harbors large number of metabolic genes, resulting in expansion of the multiple metabolic pathways [43]. Ghosh and co-workers reported Rhodococcus imtechiensis strain RKJ300 with the ability to degrade 4-nitrophenol, 2, 4-dinitro-phenol as well as their chloro substituted analogue 2-chloro-4-nitrophenol [42]. Based on the above rationale, the screening for the degradation of 2-C-4-NA was performed by using previous lab isolates known for the degradation of nitroaromatic compounds. The 2-C-4-NA degrading abilities were examined by analyzing the increase in growth and also by measuring the release of NO22,
Cl2, and NH
3in the medium. Results from primary screening,
five strains belonging to the genusRhodococcusshowed the apparent increase of bacterial cell mass as well as the release of NO22, Cl2
ions, and NH3. All these five isolates have been previously
characterized for the degradation of chloro and nitro substituted aromatic compounds. Interestingly, the strain MB-P1 showed the faster degradation of 2-C-4-NA, hence it was selected for the detailed study. During the growth study, the rapid disappearance of 2-C-4-NA was accompanied by the release of NO22ions and
NH3 followed by the release of Cl2 ions in the media. The
amounts of chloride and nitrite ions accumulated in growth media were stoichiometric; however, very limited amounts of NH3was
released. There was no accumulation of intermediates of 2-C-4-NA degradation in the growth study. Based on the growth study it could be argued that 2-C-4-NA was completely mineralized and NH3is the preferred nitrogen source for the growth of strain
MB-P1. The above results are in close agreement with the results of 5-nitroanthranilic acid degradation by Bradyrhizobium sp. strain JS329 [44]. Although, the growth study did not show any accumulation of metabolites, however, the resting cell studies showed the formation of 4-A-3-CP as the first intermediate, which subsequently disappeared with the formation of 6-CHQ as a second metabolite. Results from elucidation of the catabolic pathway in 2-C-4-NA degradation by strain MB-P1 clearly demonstrated identification of 4-A-3-CP and 6-CHQ as major intermediates. There are two ways for the removal of nitro group from the chloronitrobenzenes or chloronitrophenols out of which one is oxidative and another one is reductive reaction mechanism. In the oxidative reaction the nitro group is removed in the form of NO22ions as a results of hydroxylation, while a partial reduction
of nitro group into hydroxylamine or amino group takes place in reductive reaction [42,45–49]. Results showing transformation of 2-C-4-NA to 4-A-3-CP for initiation of 2-C-4-NA degradation by strain MB-P1 suggest the involvement of oxygenation reaction by a putative monooxygenase. The monooxygenase enzyme assays from the crude cell extracts confirm the involvement of flavin-dependent monooxygenase. The flavin-flavin-dependent monooxygen-ase reaction is very common for aerobic microbial degradation of nitro or chloro containing aromatic compounds such as 2-chloro-4-nitrophenol, 2-chloro-4-nitrophenol, 2,4-dichlorophenol and 4-chloro-phenol by aerobic microorganisms [42,50–51]. In the proposed pathway, conversion of 4-A-3-CP into 6-CHQ is expected to be catalyzed by an enzyme capable of removing amino group via oxidative reaction. Mostly the removal of amino group from the benzene ring occurs by the action of catechol dioxygenase [44]. Such catalytic reaction mechanisms are widely distributed among the microorganisms capable of degrading anilines and chloroani-lines [22,33,40,52–55].
These microorganisms utilize aniline as the sole carbon, nitrogen, and energy source and degradation occurs via the formation of catechol metabolite with the action of catechol dioxygenase [22,56–60]. Schukat et al. [61] reported the co-metabolic degradation of chloroanilines byRhodococcussp. An117 via the formation of catechol as terminal intermediate. Similarly, Zeyer et al. [62] reported utilization of chloroaniline byMoraxella
sp. strain G occurs byortho-pathway. Thus, the enzyme which is involved in the second step degradation of 2-C-4-NA i. e. conversion of 4-A-3-CP to 6-CHQ should be almost similar with those responsible for aniline and chloroaniline catabolism. The identified metabolite 6-CHQ from the resting cell study constitute the ring cleavage substrate during the 2-C-4-NA degradation by
Rhodococcussp. strain MB-P1 as supported by the accumulation of 6-CHQ during the ring cleavage inhibition study. The above result is in close agreement with results reported for
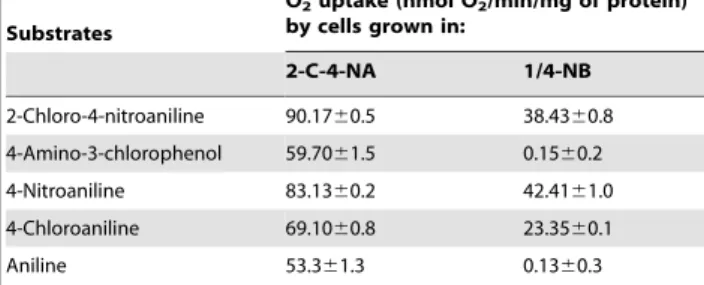
4-chlorophe-Table 1.Determination of aniline dioxygenase activity in strain MB-P1 measured by oxygen uptake by 2-C-4NA-grown versus 1/4-NB-grown cellsa.
Substrates
O2uptake (nmol O2/min/mg of protein)
by cells grown in:
2-C-4-NA 1/4-NB
2-Chloro-4-nitroaniline 90.1760.5 38.4360.8 4-Amino-3-chlorophenol 59.7061.5 0.1560.2
4-Nitroaniline 83.1360.2 42.4161.0
4-Chloroaniline 69.1060.8 23.3560.1
Aniline 53.361.3 0.1360.3
aThe reaction was carried in 1.85 ml volume air-saturated phosphate buffer
(20 mM, pH 7.2) containing substrates (40mM), and cells (0.25 mg of protein). Data represents means of at least three separate experiments.
doi:10.1371/journal.pone.0062178.t001
Figure 4. Ring cleavage inhibition studies using 2, 2-dipyridyl. (A) Treated with 2, 2-dipyridyl. (B) Untreated with 2, 2-dipyridyl. 2-C-4-NA (
N
); 4-A-3-CP (m); and 6-CHQ (#).Values are presented as arithmetic means of data obtained from experiments carried out in triplicates; error bars represent standard deviation.nol degradation by Arthrobacter chlorophenolicus A6 [27]. Based on such reports as well as the results obtained during the present study, we propose the possible involvement of a similar bacterial monooxygenase in the first step of 2-C-4-NA degradation followed by aniline dioxygenase mediated reaction as the second step of degradation. Subsequent degradation of terminal intermediate 6-CHQ presumably proceeds via conventional 2, 4, 6-trichlorophe-nol degradation pathway.
Conclusion
In conclusion, we report metabolism of 2-C-4-NA byRhodococcus
sp. strain MB-P1 which was previously characterized for atrazine degradation. This is one of the first aerobic bacteria capable of degrading 2-C-4-NA as the sole carbon, nitrogen, and energy source. Strain MB-P1 degrades 2-C-NA via the formation of 4-A-3-CP and 6-CHQ as the novel intermediates. The first step of 2-C-4-NA degradation occurs by flavin-dependent monooxygenase mediated reaction with the formation of 4-A-3-CP which gets subsequently transformed to 6-CHQ via dioxygenation reaction. The lack of induction period and accumulation of 4-A-3-CP metabolite during the induction study of 2-C-4-NA degradation by strain MB-P1 confirmed the constitutive activity of first enzyme i. e. flavin-dependent monooxygenase. The above results were also
supported by the results of oxygen uptake. However, the second enzyme responsible for the conversion of 4-A-3-CP to 6-CHQ is inducible in nature as confirmed from the oxygen uptake and induction studies. Strain MB-P1 could be used as a model system for studies focusing on this important transformation reaction for the degradation of chloro and nitro group containing anilines. Strain MB-P1 could also be used as an important model system for studies on biochemical and molecular evolution of microbial degradation of 2-C-4-NA. From application point of view, strain MB-P1 could be potentially used for bioremediation of ecological niches contaminated with 2-C-4-NA. The molecular components involved in the pathway of 2-C-4-NA degradation by strain MB-P1 are yet to be characterized.
Acknowledgments
FK, DP and SV acknowledge their research fellowship awards from CSIR, India. This is the IMTECH communication number 094/2012.
Author Contributions
Conceived and designed the experiments: SSC FK. Performed the experiments: FK DP SV. Analyzed the data: FK SSC. Wrote the paper: SSC FK.
References
1. Hampercht R, Westerkamp A (2000) Disperse Dyes. Ullmann’s Encylopedia of Industrial Chemistry (7th
Edition). NY, NY: John Wiley and Sons. Published online: 15 Sept 2000.
2. Lewis RJ, Sr. Hawley’s Condensed Chemical Dictionary (14thEdition) (2002). John Wiley and Sons, Inc. New York, NY P. 255.
3. Schnorbach HJ, Matthaei HD, Muller F (2008) Molluskicides. Ullmann’s Encylopedia of Industrial Chemistry (7thEdition). NY, John Wiley and Sons. Published online: 15 Oct 2008.
4. Graebing PW, Chib JS, Hubert TD, Gingerich WH (2004) Aqueous photolysis of niclosamide. J Agric Food Chem 52: 870–878.
5. Espinosa-Aquirre JJ, Reyes RE, Cortinas de Nava C (1991) Mutagenic activity of 2-chloro-4-nitroaniline and 5-chlorosalicyclic acid inSalmonella typhimurium: two possible metabolites of niclosamide. Mutat Res 264: 139–145.
6. Borcherding J, Wolf J (2001) The influence of suspended particles on the acute toxicity of 2-chloro-4-nitroaniline, cadmium, and pentachlorophenol on the
valve movement response of the zebra mussel (Dreissena polymorpha). Arch Environ Contam Toxicol 40: 497–504.
7. Cottalasso D, Pronzato MA, Domenicotti C, Barisione G, Fontana L, et al. (1991) Toxicity of 4-chloro-2-nitroaniline and 2-chloro-4-nitroaniline to isolated rat hepatocytes. Med Lav 82: 253–260.
8. Altschuh J, Bruggemann R, Santl H, Eichinger G, Piringer OG (1999) Henry’s law constants for a diverse set of organic chemicals: Experimental determination and comparison of estimation methods. Chemosphere 39: 1871–1887. 9. Lyman WJ, Reehl WJ, Roseblatt DH (1990) Handbook of Chemical Property
Estimation Methods: Environmental behaviour of organic compounds. Wash-ington, DC: American Chemical Society. pp. 7–4, 7–5, 8–12.
10. Struijs J, Stoltenkamp J (1986) Ultimate biodegradation of 2-, 3- and 4-nitrotoluene. Sci Total Environ 57: 161–218.
11. Canton JH, Slooff W, Kool HJ, Struys J, Pouw ThJM, et al. (1985) Toxicity, biodegradability, and accumulation of a number of Cl/N-containing compounds Figure 5. Proposed metabolic pathway for aerobic degradation of 2-C-4-NA byRhodococcussp. strain MB-P1.According to the results obtained presented here, 4-A-3-CP and 6-CHQ are identified as major intermediates during degradation of 2-C-4-NA.
for classification and establishing water quality criteria. Requl Toxicol Pharmacol 5: 123–131.
12. Saupe A (1999) High-rate biodegradation of 3- and 4-nitroaniline. Chemosphere 39: 2325–2346.
13. Qureshi A, Verma V, Kapley A, Purohit HJ (2007) Degradation of 4-nitroaniline byStenotrophomonasstrain HPC 135. Internat Biodeterio Biodegrad 60: 215–218.
14. Vangnai AS, Petchkroh W (2007) Biodegradation of 4-chloroaniline by bacteria enriched from soil. FEMS Microbiol Lett 268: 209–216.
15. Khalid A, Arshad M, Crowley DE (2009) Biodegradation of pure and mixed bacterial cultures from removal of 4-nitroaniline from textile dye wastewater. Water Res 43: 1110–1116.
16. Zhang LL, He D, Chen JM, Liu Y (2010) Biodegradtion of 2-chloroaniline, 3-chloroaniline, and 4-chloroaniline by a novel strainDelftia tsuruhatensis H1. J Hazard Mater 179: 875–882.
17. Hongsawat P, Vangnai AS (2011) Biodegradation pathways of chloroanilines by
Acinobacter baylyistrain GFJ2. J Hazard Mater 186: 1300–1307.
18. Yao XF, Khan F, Pandey R, Pandey J, Mourant RG, et al. (2011) Degradation of dichloroaniline isomers by a newly isolated strain,Bacillus megateriumIMT21. Microbiology 157: 721–726.
19. Fazlurrahman, Batra M, Pandey J, Suri CR, Jain RK (2009) Isolation and characterization of an atrazine-degradingRhodococcussp. strain MB-P1 from contaminated soil. Lett Appl Microbiol 49: 721–729.
20. Shettigar M, Pearce S, Pandey R, Khan F, Dorrian SJ, et al. (2012) Cloning of a novel chloronicotinic acid chlorohydrolase from the newly isolated 6-chloronicotinic acid mineralizingBradyrhizobiaceaestrain SG-6C. PLoS One 7: e51162.
21. Kadiyala V, Spain JC (1998) A two-component monooxygenase catalyses both hydroxylation of p-nitrophenol and oxidative release of nitrite from 4-nitrocatechol inBacillus sphericusJS905. Appl Environ Microbiol 64: 2479–2484. 22. Liu Z, Yang H, Huang Z, Zhou P, Liu S-J (2002) Degradation of aniline by newly isolated, extremely aniline-tolerant Delfitia sp. AN3. Appl Microbiol Biotechnol 58: 679–682.
23. White GF, Snape JR, Nicklin S (1996) Biodegradation of glycerol trinitrate and pentaerythritol tetranitrate byAgrobacterium radiobacter. Appl Environ Microbiol 62: 637–642.
24. Manickam N, Mau M, Schlomann M (2006) Characterization of the novel HCH-degrading strain,Microbacteriumsp. ITRC. Appl Microbiol Biotechnol 69: 580–588.
25. Tong C, Guo Y, Liu W (2010) Simlutaneous determination of five nitroaniline and dinitroaniline isomers in wastewaters by solid-phase extraction and high-performance liquid chromatography with ultraviolet detection. Chemosphere 81: 430–435.
26. Apajalathi JH, Salkinoja-Salonen MS (1987) Complete dechlorination of tetrachlorohydroquinone by cell extracts of pentachlorophenol-induced Rhodo-coccus chlorophenolicus. J Bacteriol 169: 5125–5130.
27. Nordin K, Unell M, Jansson JK (2005) Novel 4-chlorophenol degradation gene cluster and degradation route via hydorxyquinol inArthrobacter chlorophenolicusA6. Appl Environ Microbiol 71: 6538–6544.
28. Li D-Y, Eberspacher J, Wagner B, Kuntzer J, Lingens F (1991) Degradation of 2, 4, 6-trichlorophenol byAzotobactersp. strain GP1. Appl Environ Microbiol 57: 1920–1928.
29. Zaborina O, Latus M, Eberspacher J, Golovleva A, Lingens F (1995) Purification and characterization of 6-chlorohydroxyquinol 1, 2-dioxygenase from Streptomyces rochei 303: comparison with an analogous enzyme from
Azotobactersp. strain GP1. J Bacteriol 177: 229–234.
30. Golovleva LA, Zaborina O, Pertsova R, Baskunov B, Schurukhin J, et al. (1992) Degradation of polychlorinated phenols byStreptomyces rochei303. Biodegradation 2: 201–208.
31. Padilla L, Matus V, Zenteno P, Gonzalez B (2000) Degradation of 2, 4, 6-trichlorophenol via chlorohydroxyquinol in Ralstonia eutropha JMP134 and JMP222. J Basic Microbiol 40: 243–249.
32. Sanchez MA, Gonzalez B (2007) Genetic characterization of 2, 4, 6-trichlorophenol degradation in Cupriavidus necator JMP134. Appl Environ Microbial 73: 201–208.
33. Liang Q, Takeo M, Chen M, Zhang W, Xu Y, et al. (2005) Chromosomal-encoded gene cluster for the metabolic pathway that converts aniline to TCA cycle intermediates inDelftia tsuruhatensisAD9. Microbiology 151: 3435–3446. 34. Shin KA, Spain JC (2009) Pathway and evolutionary implication of
diphenylamine biodegradation byBurkholderiasp. strain JS667. Appl Environ microbiol 75: 2694–2704.
35. Kitagawa W, Kimura N, Y Kamagata Y (2004) A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium,Rhodococcus opacus
SAO101. J Bacteriol 186: 4894–4902.
36. Takeo M, Murakami M, Niihara S, Yamamoto K Nishimura M, et al. (2008) Mehcanism of 4-nitrophenol oxidation inRhodococcussp. strain PN1:
Charac-terization of the two-component 4-nitrophenol hydroxylase and regulation of its expression. J Bacteriol 190: 7367–7374.
37. Wei M, Zhang JJ, Liu H, Zhou NY (2010)para-nitrophenol 4-monooxygenase and hydroxyquinol 1, 2-dioxygenase catalyze sequential transformation of 4-nitrocatechol inPseudomonassp. strain WBC-3. Biodegradation 21: 915–921. 38. Lovley DR (2003) Cleaning up with genomics: applying molecular biology to
bioremediation. Nat Rev Microbiol 1: 35–44.
39. Paul D, Pandey G, Pandey J, Jain RK (2005) Accessing microbial diversity for bioremediation and environmental restoration. Trends Biotechnol 23: 135–142. 40. Boon N, Goris J, De Vos P, Verstraete W, Top EM (2001) Genetic diversity among 3-chloroaniline and aniline degrading strains ofComamonadaceae. Appl Environ Microbiol 67: 1107–1115.
41. Sun JH, Sun SP, Fan MH, Guo HQ, Qiao LP, et al. (2007) A kinetic study on the degradation ofp-nitroaniline by Fenton oxidation process. J Hazard Mater 148: 172–177.
42. Ghosh A, Khurana M, Chauhan A, Chakraborti AK, Jain RK (2010) Degradation of 4-nitrophenol, 2-chloro-4-nitrophenol, and 2, 4-dinitrophenol byRhodococcus imtechensisstrain RKJ300. Environ Sci Technol 44: 1069–1077. 43. Larkin MJ, Kulakov LA, Allen CCR (2005) Biodegradation andRhodococcus
-master of catabolic versality. Curr Opin Biotechnol 16: 282–290.
44. Qu Y, Spain JC (2010) Biodegradation of 5-nitroanthranilic acid by
Bradyrhizobiumsp. strain JS329. Appl Environ Microbiol 76: 1417–1422. 45. Arora PK, Srivastava A, Singh VP (2010) Application of monooxygenase in
dehalogenation, desulphurization, denitrification and hydroxylation of aromatic compounds. J Bioremed Biodegrad 1: 112.
46. Arora PK, Jain RK (2012) Metabolism of 2-chloro-4-nitrophenol in a gram-negative bacterium,Burkholderiasp. RKJ800. PloS ONE 7(6): e38676. 47. Beunink J, Rehm HJ (1990) Coupled reductive and oxidative degradation of
4-chloro-2-nitrophenol by a co-immobilized mixed culture system. Appl Microbiol Biotechnol 34: 108–115.
48. Katsivela E, Wray V, Pieper DH, Wittich RM (1999) Initial reactions in the biodegradation of 1-chloro-4-nitrobenzene by a newly isolated bacterium, strain LW1. Appl Environ Microbiol 65: 1405–1412.
49. Wu JF, Jiang CY, Wang BJ, Ma YF, Liu ZP, et al. (2006) Novel partial reductive pathway for 4-chloronitrobenzene and nitrobenzene degradation inComamonas
sp. strain CNB-1. Appl Environ Microbiol 72: 1759–1765.
50. Perry LL, Zylstra GJ (2007) Cloning of a gene cluster involved in the catabolism ofp-nitrophenol byArthrobactersp. strain JS443 and characterization of thep -nitrophenol monooxygenase. J Bacteriol 189: 7563–7572.
51. Nishino SF, Spain JC (2004) Catabolism of nitroaromatic compounds, p. 575-608. In J.L. Ramos (ed.),Pseudomonas, vol III. Biosynthesis of macromolecules and molecular metabolism. Kluwer Academic/Plenum Publishers, New York, NY
52. Aoki K, Shinke R, Nishira H (1983) Metabolism of aniline by Rhodococcus erythropolisAN-13. Agric Biol Chem 47: 1611–1616.
53. Murakami S, Nakanishi Y, Shinke R, Aoki R (1991) Catechol 1, 2-dioxygenase isozymes in soil bacteria metabolizing aromatic compounds. Soil Biol Biochem 23: 815–819.
54. Kahng HY, Kukor JJ, Oh KH (2000) Characterization of strain HY99, a novel microorganism capable of aerobic and anaerobic degradation of aniline. FEMS Microbiol Lett 190: 215–221.
55. Travkin VM, Solyanikova IP, Rietjens IM, Vervoort J, van Berkel WJ, et al. (2003) Degradation of 3, 4-dichloro and 3, 4-difluoroaniline by Pseudomonas fluorescens26-K. J Environ Sci Health B 38: 121–132.
56. Aoki K, Konohana T, Shinke R, Nishira H (1984) Purification and characterization of catechol 1, 2-dioxygenase from aniline-assimilating Rhodo-coccus erythropolisAN-13. Agric Biol Chem 48: 2087–2095.
57. Murakami S, Kodama N, Shinke R, Aoki K (1997) Classification of catechol 1, dioxygenase family: sequence analysis of a gene for the catechol 1, 2-dioxygenase showing high specificity for methylcatechols from gram+aniline assimilatingRhodococcus erythropolisAN-13. Gene 185: 49–54.
58. Strachan PD, Freer AA, Fewson CA (1998) Purification and characterization of catechol 1, 2-dioxygenase from Rhodococcus rhodochrous NCIMB 13259 and cloning and sequencing of its catA gene. Bicohem J 333: 741–747.
59. Kutty R, Purohit HJ, Khanna P (2000) Isolation and characterization of
Pseudomonassp. strain PH1 utilizingmeta-aminophenol. Can J Microbiol 46: 211– 217.
60. Matsumura E, Sakai M, Hayashi K, Muakami S, Takenaka S, et al. (2006) Constitutive expression of catABC gene in the aniline-assimilating bacterium
Rhodococcusspecies AN-22: production, purification, characterization and gene analysis of CatA, CatB and CatC. Biochem J 393: 219–226.
61. Schukat B, Janke D, Krebs D, Fritsche W (1983) Cometabolic degradation of 2-and 3-chloroaniline because of glucose metabolism byRhodococcussp. An117. Curr Microbiol 9: 81–86.